Abstract
In this study, the complete mitogenome sequence of Vespula flaviceps (Hymenoptera, Vespidae) has been sequenced. The mitogenome, 17,489 bp, includes 13 protein-coding, 22 transfer RNAs, two ribosomal RNAs genes and a noncoding D-loop region. The D-loop of 1302 bp length is located between rRNA-S and tRNATyr. The overall base composition of V. flaviceps is 39.34% for A, 13.62% for C, 40.99% for T, and 6.05% for G, with a high AT bias of 80.33%. This mitogenome data might be useful for further phylogeography analyses and other related studies in Hymenoptera.
Keywords:
Vespula flaviceps (Hymenoptera, Vespidae, Vespula) is a species of social wasp, mainly distributed in Eastern and Southern Asia. This wasp is popular as food in Southern China, Central Japan, and Korea, and its larvae are considered as a special delicacy. Elucidating the structure of V. flaviceps mitogenome is important for understanding its diversity and evolution.
The specimen of V. flaviceps was obtained from Kunming, Yunnan, P.R. China and kept in the Insect Collection of Research Institute of Resource Insects, Chinese Academy of Forestry. Total genomic DNA was extracted from its chest muscle with SDS-phenol chloroform method. Sequencing work of the complete mitogenome of V. flaviceps was performed by Illumina Nextseq500 in Beijing Microread Genetics Co., Ltd., with a total data volume 4G (150 bp Reads), and high-quality reads were assembled from scratch using IDBA-UD and SPAdes (Gurevich et al. Citation2013).
The complete mitogenome of V. flaviceps was found to be bigger than most other invertebrates. The gene order and orientation are also different from the most common type suggested as ancestral for insects (Boore et al. 1998; Taanman Citation1999). It was 17,489 bp in size (GenBank accession number MK248830), including 13 typical invertebrate protein-coding genes (PCGs), 22 transfer RNA genes, two ribosomal RNA genes and a noncoding control region (D-loop). The A + T content of the whole V. flaviceps mitogenome is 80.33%, showing an obvious AT mutation bias (Eyre-Walker Citation1997). The D-loop region exhibits the highest A + T content (88.33%) in the V. flaviceps mitogenome. This region has been shown to harbor the origin sites for the transcription and replication for both strands of insect mitogenomes (Yukuhiro et al. Citation2002).
All the 13 PCGs use standard ATN (Met) start codon (ATG in ND2, ATP6, COX3, ND4, ND6, CytB; ATT in COX1, COX2, ATP8; ATA in ND3, ND5, ND4L, ND1) All the PCGs, except COX3, had the common stop codon TAA, whereas COX3 terminated with incomplete stop codon T. Similar cases could be found in other insect mitogenomes (Yin et al. Citation2012). It has been demonstrated that the incomplete stop codons could produce functional stop codons ‘TAA’ in polycistronic transcription cleavage and polyadenylation processes by the addition of 3’A residues (Ojala et al. Citation1981).
All the tRNAs except tRNASer(AGN) could be folded into the typical cloverleaf secondary structures. The unusual tRNASer(AGN) lacks dihydrouridine (DHU) arm. The rRNA-S gene is located between tRNAVal and the D-loop region, while rRNA-L is located between ND1 and tRNAVal. In other insects, the rRNA-L gene is usually located between tRNALeu (CUN) and tRNAVal (Boore et al. Citation1998; Kim et al. Citation2009), this demonstrated that in V. flaviceps mitogenome the positions of ND1 and tRNALeu (CUN) were interchanged.
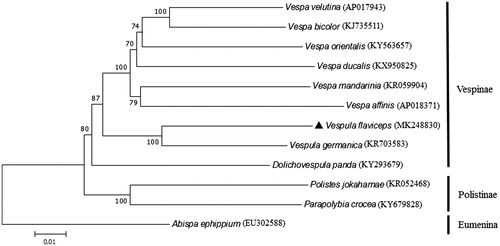
Based on the concatenated amino acid sequences of 13 PCGs, the neighbour-joining method was used to construct the phylogenetic relationship of V. flaviceps with 11 other wasps (). The result showed that V. flaviceps is closely related to V. germanica, which is in accordance with the traditional morphological classification (Carpenter and Nguyen Citation2003).
Disclosure statement
The authors declare no competing materials in the preparation and execution of this manuscript. The authors are responsible for the content and writing of this paper.
Additional information
Funding
References
- Boore JL, Lavrov DV, Brown WM. 1998. Gene translocation links insects and crustaceans. Nature. 392:667–668.
- Carpenter JM, Nguyen LPT. 2003. Keys to the genera of social wasps of South-East Asia (Hymenoptera: Vespidae). Entomol Sci. 6: 183–192.
- Eyre-Walker A. 1997. Differentiating between selection and mutation bias. Genetics. 147:1983–1987.
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29: 1072–1075.
- Kim MI, Baek JY, Kim MJ, Jeong HC, Kim KG, Bae CH, Han YS, Jin BR, Kim I. 2009. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol Cells. 28:347–363.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Taanman JW. 1999. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta. 1410:103–123.
- Yin H, Zhi Y, Jiang H, Wang P, Yin X, Zhang D. 2012. The complete mitochondrial genome of Gomphocerus tibetanus Uvarov, 1935 (Orthoptera: Acrididae: Gomphocerinae). Gene. 494:214–218.
- Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y. 2002. Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori. Mol Biol Evol. 19:1385–1389.