Abstract
The complete mitochondrial genome of Coelorinchus multispinulosus was determined using the MiSeq platform. The mitogenome of C. multispinulosus (15,937 bp) contained 13 protein-coding genes, 22 tRNAs, two ribosomal RNAs (12S and 16S), and a control region (D-Loop). The G + C content of its mitogenome was 44.69%, which was lower than A + T contents (55.31%). Unusual start codon (GTG) was identified in three protein-coding genes including COX1, ND2, and ND3. Incomplete stop codons (TA–/T–) were shown in six genes including COX1, COX2, COX3, ND3, ND4, and ATP6. Based on the phylogenetic tree with the currently reported mitogenomes in the family Macrouridae, C. multispinulosus was most closely related to Coelorinchus kishinouyei with 85% nucleotide sequence identity.
As the largest genus in the family Macrouridae, about 120 species are currently known in the coelorinchus and many of which are awaiting further description (Iwamoto and Williams Citation1999; Iwamoto Citation2008). The fish in the genus generally inhabit on the continental shelves and slope in the tropical and subtropical ocean and Southeast Asia is known to be the center of its diversity (Iwamoto Citation2008). Despite the large species numbers in the genus, only one mitogenome sequence in the coelorinchus is currently reported. We here report the complete mitochondrial genome of the Spearnose grenadier, Coelorinchus multispinulosus, which was collected from Korean water.
The specimen was collected from a regular EEZ fish survey (32°03'28.3''N, 125°05'58.6''E) by National Institute of Fisheries Science (NIFS) in 2013. The specimen and its DNA was stored in Pukyong National University. Identification of the specimen was confirmed by both morphological characteristics and by its 100% sequence identity in COI region (GenBank number: DQ648448). Based on the COI region, C. multispinulosus was most closely related to Coelorinchus formosanus with 93% sequence identity (GenBank number: KP267634). The complete mitochondrial genome sequence of C. multispinulosus was determined by the bioinformatic assembly of the high-throughput sequence (HTS) reads. The mitochondrial DNA extracted using the commercial kit (Abcam, UK) was further fragmented into smaller sizes (∼350 bp) by Covaris M220 Focused-Ultrasonicator (Covaris Inc., Woburn, MA, USA). A library was constructed using TruSeq® RNA library preparation kit V2 (Illumina Inc., San Diego, CA, USA), and its quality and the quantity was analyzed using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The MiSeq sequencer (2 × 300 bp pair ends) was used for the sequencing (Illumina Inc., San Diego, CA, USA). Geneious® 11.0.2 (Kearse et al. Citation2012) was used for the mitogenome assembly.
The complete mitochondrial genome of C. multispinulosus (GenBank Number: MK386892) was 15,937 bp in length, which harbors 13 protein-coding genes, 22 tRNAs, two ribosomal RNAs (12S and 16S), and a control region (D-Loop). Its G + C content was 44.69%, which was lower than A + T contents (55.31%). Eight genes were located on the light (L) strand, whereas the remaining 29 genes were on the heavy (H) strand. Except for ND6, the other 12 protein-coding genes were located on H strand. Ten protein-coding genes started with common ATG, whereas COX1, ND2, and ND3 showed unusual GTG as the start codon. The incomplete stop codons (TA–/T–) were identified in six genes including COX1, COX2, COX3, ND3, ND4, and ATP6, whereas unusual stop codons were found in ND6 (AGG) and Cytb (AGA).
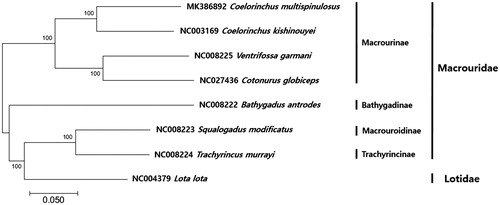
The phylogenetic analysis of C. multispinulosus with the currently reported mitogenomes in the family Macrouridae was constructed using MEGA7.0 program with minimum evolutionary (ME) algorithm (Kumar et al. Citation2016). The result showed that C. multispinulosus was most closely related to the Coelorinchus kishinouyei (85%), followed by Ventrifossa garmani (82%) and Cetonurus globiceps (77%) ().
Figure 1. Phylogenetic tree of Coelorinchus multispinulosus in family Macrouridae. Phylogenetic tree of C. multispinulosus complete genome was constructed using MEGA7 software with Minimum Evolution (ME) algorithm with 1000 bootstrap replications. GenBank Accession numbers were shown followed by each species scientific name.
Disclosure statement
The authors reported that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Iwamoto T. 2008. Two new Australian grenadiers of the Coelorinchus fasciatus species group (Macrouridae: Gadiformes: Teleostei). Proc Calif Acad Sci (Ser 4). 59:133–146.
- Iwamoto T, Williams A. 1999. Grenadiers (Pisces, Gadiformes) from the continental slope of western and northwestern Australia.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
