Abstract
We sequenced the chloroplast genome of Skeletonema pseudocostatum clone SZCZ 1832 from Algeria. The genome is 127,013 bp long. The inverted repeat region is 18,240 bp long and the order of the genes it encodes appeared as highly conserved when compared with other Thalassiosirales. The maximum likelihood phylogeny also evidenced the belonging of S. pseudocostatum within the Thalassiosirales. This chloroplastic genome is so far the first made available for the cosmopolitan and ecologically important genus Skeletonema.
Keywords:
Skeletonema is a genus of centric, planktonic diatoms that belongs to the Thalassiosirales. Members of this genus are distributed worldwide and can produce massive blooms (Sarno et al. Citation2005). Some species are commonly used in aquaculture for feeding bivalves and crustaceans (Brown Citation2002; Sirakov et al. Citation2015) and can also serve as models in ecotoxicological studies (e.g. Chen et al. Citation2018; Zhu et al. Citation2019). Despite the numerous studies devoted to Skeletonema, no chloroplast genome sequence is currently available for any species. In the framework of a collaborative public-private partnership between the University of Oran 1 (Algeria), the ESSBO (Algeria) and the WiraGen company (Algeria) aiming at developing bioprospecting, molecular taxonomy and blue biotechnologies in North Africa, we have undertaken several sampling campaigns of the coastal waters of Algeria. During one of these campaigns, a strain of Skeletonema pseudocostatum Medlin (Medlin et al. Citation1991) was sampled near Cap Falcon (35.769671N, 0.799587W), isolated, and maintained in culture. Mounted scanning electron microscope stubs and permanent slides with cleaned frustules of this S. pseudocostatum isolate are available from the University of Szczecin (Poland) under the accession SZCZ 1832 and live cultures can be obtained from both the University of Oran 1 and the University of Szczecin. DNA was extracted following the procedure described by Doyle and Doyle (Citation1990) and sequenced on the BGISEQ-500 platform at the Beijing Genomic Institute. A total of 60 million 100 bp reads were assembled using SPAdes 3.12.0 (Bankevich et al. Citation2012), and the contig corresponding to the chloroplast genome was annotated using the findORF tools (Gagnon Citation2004).
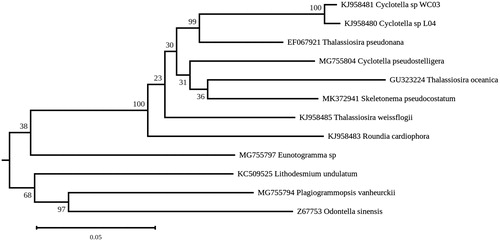
The Skeletonema pseudocostatum chloroplast genome is 127,013 bp in size and contains a large inverted repeat sequence of 18,240 bp (GenBank: MK372941) that encodes 21 genes ordered as follows: trnP(ugg), ycf89, rrs, trnI(gau), trnA(ugc), rrl, rrf, psbY, rpl32, trnL(uag), rbcR, rpl21, rpl27, secA, rpl34, ycf46, ccs1, psbA, ycf35, psaC and ccsA. The gene content and gene organization of the inverted repeat are similar to those observed in other members of Thalassiosirales (Sabir et al. Citation2014). The rbcL gene of the analyzed strain proved to be 100% identical to that reported for the S. pseudocostatum CCAP1077/7 strain (GenBank: DQ514819), which was also isolated from the North African shores (https://www.ccap.ac.uk/strain_info.php?Strain_No=1077/7). Using RAxML Version 8 (Stamatakis Citation2014), a maximum likelihood phylogeny was inferred from concatenated gene sequences derived from photosystem I and photosystem II genes (psa and psb genes, respectively) of S. pseudocostatumand other diatoms whose chloroplast genome is currently available (subtree displayed in ). The results unambiguously showed that S. pseudocostatum clusters with other members of the Thalassiosirales. Similar genomic studies on other diatoms from Algeria are underway.
Acknowledgements
We would like to thank the WiraGen company (https://www.genewir.com/) for the technical support and advices. We also thank Ming Lei from BGI Europe for the help with the sequencing process.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Brown MR. 2002. Nutritional value of microalgae for aquaculture. In: Cruz-Suárez, LE., Ricque-Marie D, TapiaSalazar M, Gaxiola-Cortés MG, Simoes N., editors. Avances en Nutrición Acuícola VI. Memorias del VI Simposium Internacional de Nutrición Acuícola. 3 al 6 de septiembre del 2002. Cancún, Quintana Roo, México: Universidad Autónoma de Nuevo León. Monterrey, N.L.
- Chen X, Zhang C, Tan L, Wang J. 2018. Toxicity of Co nanoparticles on three species of marine microalgae. Environ Pollut. 236:454–461.
- Doyle JJ, Doyle JL. 1990. Isolation of plant DNA from fresh tissue. Focus. 12:13–15.
- Gagnon J. 2004. Création d’outils pour l’automatisation d’analyses phylogénétiques de génomes d’organites [Development of bioinformatic tools for the phylogenetic analyzes of organellar genomes] [M.Sc. dissertation]. Québec city: University of Lava.
- Medlin LK, Elwood HJ, Stickel S, Sogin ML. 1991. Morphological and genetic variation within the diatom Skeletonema costatum (Bacillariophyta): evidence for a new species, Skeletonema pseudocostatum. J Phycol. 27:514–524.
- Sabir JS, Yu M, Ashworth MP, Baeshen NA, Baeshen MN, Bahieldin A, Theriot EC, Jansen RK. 2014. Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS One. 9:e107854.
- Sarno D, Kooistra WH, Medlin LK, Percopo I, Zingone A. 2005. Diversity in the genus Skeletonema (Bacillariophyceae). II. An assessment of the taxonomy od S. costatum like species with the description of four new species. J Phycol. 41:151–176.
- Sirakov I, Velichkova K, Stoyanova S, Staykov Y. 2015. The importance of microalgae for aquaculture industry Review. Int J Fish Aquat Stud. 2:81–84.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zhu ZL, Wang SC, Zhao FF, Wang SG, Liu FF, Liu GZ. 2019. Joint toxicity of microplastics with triclosan to marine microalgae Skeletonema costatum. Environ Pollut. 246:509–517.