Abstract
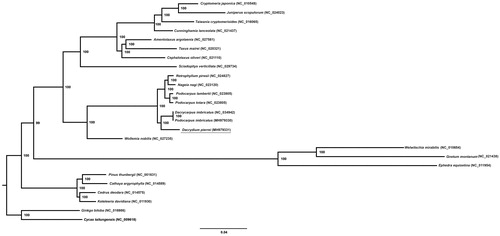
In this study, the complete chloroplast genome of Dacrydium pierrei was reported. The results showed that the whole genome was 131,671 bp in length. The genome contains a total of 118 encoding genes, including 82 protein-coding genes, 32 tRNA genes, and 4 rRNA genes. Fourteen genes contain one intron and ycf3 gene contains two introns. The GC content of the entire genome is 37.25%. The maximum likelihood phylogenetic analysis based on 59 chloroplast genes from 25 gymnosperm taxa showed that D. pierrei was sister to Podocarpus imbricatus.
Chloroplast (cp) genomes usually contain highly conserved structure, including a large single copy region and a small single copy region separated by two inverted repeat sequences (IR). However, the typical IR was lost in all conifer chloroplast genomes (Raubeson and Jansen Citation1992). The chloroplast genomes from conifers offer a valuable resource for studying dynamic evolutionary process of IR.
Dacrydium pierrei belongs to the family of Podocarpaceae and is scattered in Oceania, South America, Indo-China, and Southeast Asia. The wood of D. pierrei has been widely used for high-grade architectural work and handcrafting. However, D. pierrei has been severely destroyed by excessive deforestation, leading to a significant decrease in natural forest resources (Yang et al. Citation2008). This species is listed as a third-class endangered plant in China (Keppel et al. Citation2011).
The fresh leaves of D. pierrei were obtained from South China Botanical Garden, Chinese Academy of Sciences. The voucher specimens of D. pierrei were deposited in Shaanxi Xueqian Normal University. DNA was extracted using CTAB-based method (Gawel and Jarret Citation1991). The whole-genome sequencing was carried out by the Illumina Hiseq2500 Platform (Illumina Inc., San Diego, CA). We assembled the complete chloroplast genome sequence of D. pierrei by NOVOPlasty software (Dierckxsens et al. Citation2017). DOGMA and tRNAscan-SE were used to annotate protein-coding gene and tRNA gene, respectively (Wyman et al. Citation2004; Schattner et al. Citation2005). The complete chloroplast genome sequence of D. pierrei was submitted to GenBank under the accession number MH979331.
The total size of complete cp genome is 131,671 bp in length with 74,148 bp (56.31%) coding region. Like other conifers cp genomes, D. pierrei has no typical inverted repeat sequence. The overall GC content of the genome is 37.25% and this value in the protein-coding region, tRNA gene, rRNA gene, intron and intergenic region is 38.20, 53.25, 53.89, 37.54 and 33.62%, respectively. A total of 118 genes were successfully annotated, including 82 protein-coding genes, 32 tRNA genes, and 4 rRNA genes. The gene density was estimated to be 0.90 genes/kb. Among the 118 genes, eight protein-coding and six tRNA genes are single-intron genes except for ycf3 with two introns. Two copies of trnN-GUU genes were identified. While rps16 and the intron in both clpP and rpoC1 were lost. The phylogenetic position of D. pierrei was inferred by RAxML v8.1.x using 53 protein-coding and four rRNA genes from 25 gymnosperm taxa. The tree strongly supported the sister relationship between D. pierrei and Podocarpus imbricatus (). However, the Podocarpus lambertii, Podocarpus totara, P. imbricatus (also called Dacrycarpus imbricatus) from the same genus did not form a monophytic. More comprehensive data and methods are required to infer the phylogeny of Podocarpus. In general, the complete chloroplast genome of D. pierrei provides a valuable genomic resource for phylogenetic and evolutionary analysis of Podocarpaceae.
Acknowledgments
We thank Yi-Hua Tong in South China Botanical Garden, Chinese Academy of Sciences, for sampling materials.
Disclosure statement
No potential conflict of interest was declared by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Gawel N, Jarret R. 1991. A modified CTAB DNA extraction procedure for musa and ipomoea. Plant Mol Biol Rep. 9:262–266.
- Keppel G, Prentis P, Biffin E, Hodgskiss P, Tuisese S, Tuiwawa MV, Lowe AJ. 2011. Diversification history and hybridization of Dacrydium (Podocarpaceae) in Remote Oceania. Aust J Bot. 59:262–273.
- Raubeson LA, Jansen RK. 1992. A rare chloroplast-DNA structural mutation is shared by all conifers. Biochem Syst Ecol. 20:17–24.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang YC, Zhang WY, Lin RC, Yang XS. 2008. Study on structure and species diversity in post harvested tropical montane Rain forest Dominated by Dacridium pierrii in Bawangling, Hainan Island. Forest Res. 21:70–74.