Abstract
The complete chloroplast genome of Ranunculus cantoniensis was determined. Total chloroplast genome was 155,119 bp in lenth, displayed a typical quadripartite structure, including a large single copy (LSC) region of 84,634 bp and a small single copy region (SSC) of 18,879 bp, separated by a pair of inverted repeats (IRs) of 25,803 bp. A total of 131 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes were indentified. Among them, 15 genes have one intron each and 3 genes contain two introns. The overall GC content was 37.9%, while the corresponding values of LSC, SSC, and IR regions were 36.0, 31.1, and 43.5%, respectively. Phylogenetic relationship analysis showed that R. cantoniensis was closely related to R. macranthus.
Ranunculus cantoniensis DC. is a perennial weed of Ranunculaceae widely spread in tropical and subtropical Asia (Xu et al. Citation2013). Due to the morphological plasticity, hybridization and polyploidy, generic delimitation and infrageneric classification of Ranunculus is still under consideration (Hörandl et al. Citation2005). R. cantoniensis DC. was often confused with R. chinensis, R. silerifolius var. silerifolius, R. silerifolius var. dolicanthus and R. trigonus (Kuo et al. Citation2005). These species frequently occur in sympatry and natural hybridization occur occasionally (Li et al. Citation2014), thus they were usually treated as a species complex (Tamura Citation1978). Universal plastid markers cannot provide enough variable sites to construct the phylogeny of this species complex. Chloroplast genome-scale data have proven to be useful in resolving intractable phylogenetic relationships (Yang et al. Citation2016). In this study, we reported the complete chloroplast genome sequence of R. cantoniensis.
Total genomic DNA was extracted from fresh leaves that were collected from a single individual of R. cantoniensis in Jiujiang city (115.97E; 29.69N), Jiangxi Province, China. The voucher specimen deposited in Jiujiang University (accession number JJU180701). Illumina paired-end (PE) library was prepared and sequenced on an illumine Hiseq X Ten platform. Owing to the interference of nuclear genome, chloroplast genome related reads were sieved by mapping to the closer species R. macranthus. The chloroplast genome was assembled via SOAPdenovo (Luo et al. Citation2012) using the R. macranthus chloroplast as a reference (GenBank accession NC_008796). The finished chloroplast genome was annotated by Geneious (Kearse et al. Citation2012), coupled with manual check and adjustment.
The complete chloroplast genome of R. cantoniensis (GenBank accession MK370318) is a circular form of 155,119 bp in length with high coverage (mean 323×), which is composed of four distinct regions: one large single copy (LSC) region of 84,634 bp and one small single copy region (SSC) of 18,879 bp separated by two inverted repeats (IRs) of 25,803 bp. The genome contains a total of 131 genes, including 84 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) have one intron each, and three genes (clpP, rps12 and ycf3) contain two introns. The overall GC content is 37.9%, while the GC content of LSC, SSC, and IR regions are 36.0, 31.1, and 43.5%, respectively. The complete chloroplast genome will provide valuable molecular data for species identification and phylogenetic analysis for genus Ranunculus.
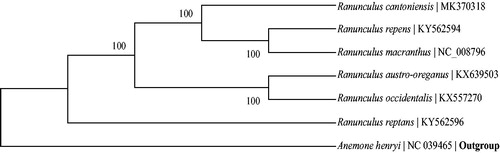
The phylogenetic tree including R. cantoniensis, five Ranunculus species and the outgroup Paeonia obovate was constructed by complete chloroplast genomes. The genome-wide alignment was carried out by software MAFFT (Katoh and Standley Citation2013). Also, the phylogenetic tree was generated by the software RaxML (Stamatakis Citation2014) with 100 bootstrap replicates. All the nodes were inferred with strong support by the ML methods. As shown in the phylogenetic tree (), R. cantoniensis is most closely related to R. macranthus.
Additional information
Funding
References
- Hörandl E, Paun O, Johansson JT, Lehnebach C, Armstrong T, Chen LX, Lockhart P. 2005. Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Mol Phylogenet Evol. 36:305–327.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kuo S, Yang T, Wang J. 2005. Revision of Ranunculus cantoniensis DC. and allied species (Ranunculaceae) in Taiwan. Taiwania. 50:209–221.
- Li T, Xu L, Liao L, Deng H, Han X. 2014. Patterns of hybridization in a multispecies hybrid zone in the Ranunculus cantoniensis complex (Ranunculaceae). Bot J Linn Soc. 174:227–239.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tamura M. 1978. Ranunculus cantoniensis group in Japan. J Geobot. 26:34–40.
- Xu L, Li T, Liao L, Deng H, Han X. 2013. Reticulate evolution in Ranunculus cantonensis polyploid complex and its allied species. Plant Syst Evol. 299:603–610.
- Yang Y, Zhou T, Duan D, Yang J, Feng L, Zhao G. 2016. Comparative analysis of the complete chloroplast genomes of five Quercus species. Front Plant Sci. 7:959.