Abstract
Konjac (Amorphophallus konjac K. Koch) is a perennial herb with high commercial and medicinal values. To better understand its genetic and genomic profiles, we sequenced and assembled the completed chloroplast genome of A. konjac. The complete chloroplast genome is 155,277 bp in length, consisting of the large (LSC, 85,412 bp) and the small single-copy region (SSC, 19,449 bp), which were separated by a pair of inverted repeat regions (IRs, 25,208 bp). It possesses 113 unique genes (80protein-coding genes, 29 tRNAs and 4rRNAs). Phylogenetic analysis suggests that A. konjac is closely related to Colocasia esculenta and Pinellia ternata.
Konjac (Amorphophallus konjac K. Koch), a perennial herb belonging to the family Araceae, is native to subtropical to tropical Southeast Asia and China (Hetterscheid and Ittenbach Citation1996). The plant has been commonly cultivated as a food crop in China, Korea, Japan, and Southeast Asia for more than 2000 years (Wootton et al. Citation1993; Long Citation1998; Bown Citation2000; Douglas et al. Citation2005). In addition, it has been traditionally used as a medicinal herb, owing to its analgesic, hemostatic, and anti-tumor activities (Chua et al. Citation2010). The complete chloroplast (cp) genome can provide new insights into the utilization of this commercially and medicinally important plant. Here, we sequenced and assembled complete cp genome of konjac using high throughput Illumina sequencing technology.
The fresh leaves of A. konjac were collected from the Botanical Garden of Kunming Institute of Botany, Chinese Academy of Sciences (25°8′21′′N, 102°44′25′′E); and the voucher specimen (LCK-20180712) was deposited at the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Total DNA was extracted from 100 mg of fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987). Ensuing nine universal pairs of primers and protocols developed by Yang et al. (Citation2014) were used to amplify the cp genome. We then sequenced the complete cp genome using Illumina Hiseq 2000 sequencing platform at BGI (Shenzhen, Guangdong, China). The raw data were filtered by NGS QC Tool Kit (Patel and Jain Citation2012) and then the high-quality reads were assembled using CLC Genomics Workbench v8.0 (CLC Bio). Contigs were mapped onto the reference cp genome of Colocasia esculenta (NC_016753) with Bowtie v2.2.6 (Langmead and Salzberg Citation2012). The assembly was edited and annotated according to the reference in Geneious V10.2 (Kearse et al. Citation2012). The validated complete cp genome of A. konjac was deposited in GenBank under the accession number KY769273.
The complete cp genome of A. konjac was 155,277 bp in length, which exhibited a typical quadripartite structure, including a pair of inverted repeated regions (IRs, 25,208 bp) separated by the large single-copy region (LSC, 85,412 bp) and the small single-copy region (SSC, 19,449 bp). It harbored 113 unique genes, including 80 protein-coding genes, 29 tRNAs, and 4 rRNAs. Among those unique genes, 11 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, rps16 and ycf68) and 6 tRNAs (trnA-UGC, trnG-UCC, trnI-GAU, trnL-UUU, trnL-UAA and trnV-UAC) possessed one intron, whereas ycf3 and clpP had two introns.
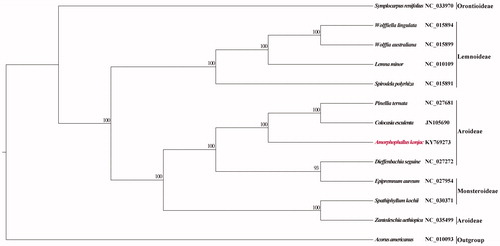
To reveal the phylogenetic position of A. konjac, a maximum likelihood tree was reconstructed using RAxML version 8.2.11 (Stamatakis Citation2014) based on 12 whole cp genomes. Acorus americanus was used to root the tree. The branch support was computed with 1000 bootstrap replicates. Phylogenetic analysis robustly revealed that A. konjac was nested in tribe Aroideae and sister to the clade including Colocasia esculenta and Pinellia ternata ().
Disclosure statement
The authors declare no potential conflict of interests.
Additional information
Funding
References
- Bown D. 2000. Aroids: plants of the Arum family. Portland: Timber Press.
- Chua M, Baldwin TC, Hocking TJ, Chan K. 2010. Traditional uses and potential health benefits of Amorphophallus konjac K. Koch ex N.E.Br. J Ethnopharmacol. 128:268–278.
- Douglas JA, Follett JM, Waller JE. 2005. Research on konjac (Amorphophallus konjac) production in New Zealand. Acta Horticul. 670:173–180.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hetterscheid W, Ittenbach S. 1996. Everything you always wanted to know about Amorphophallus, but were afraid to stick your nose into. Aroideana. 19:7–129.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Long CL. 1998. Ethnobotany of Amorphophallus of China. Acta Bot Yunnan. Suppl. X:89–92.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. Plos One. 7:e30619. doi: 10.1371/journal.pone.0030619.
- Stamatakis A. 2014. Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wootton AN, Luker-Brown M, Westcott RJ, Cheetham PSJ. 1993. The extraction of a glucomannan polysaccharide from konjac corms (elephant yam, Amorphophallus rivierii). J Sci Food Agric. 61:429–433.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Notes. 14:1024–1031.