Abstract
The complete mitochondrial genome of Nerita yoldii has been determined. The whole sequence was 15,719 bp in length and included 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes (12S rRNA and 16S rRNA). The overall base composition is A 29.5%, C 13.8%, G 21.5%, and T 35.2%. In 13 protein-coding genes, the nad2, nad5, and nad6 gene start with ATT, whereas nad4 started with ATA, the other nine genes start with ATG. For the stop codon, COX1, COX3, nad3, and atp8 end with TAG, other nine genes end with TAA. Phylogenetic tree was constructed based on 13 protein-coding genes sequences of Archaeogastropoda species, and the result showed that N. yoldii is most closely related to Clithon retropictus. These mitogenome sequence data will be useful for population genetics and phylogenetic analysis of the family Neritidae.
Nerita yoldii (Récluz Citation1841) is a common gastropod found on rocky intertidal shores along the coast of Southeast China Sea (Wang et al. 2018). It is a common economy aquatic product species. The shell size of adult varies between 17 and 20 mm, the shell has an oviform shape, and it contains many serration. A period of planktonic larvae occurs after hatching that larvae can reach shore near the Shengsi Archipelago, south of the Yangtze River Estuary, in the north, by coastal currents (Zhang et al. Citation1963; Yeung and Yang Citation2006). At present, there are only a few preliminary taxonomic and life cycle studies on N. yoldii, and there is still a lack of genetic research, which needs further elucidated.
In the present study, we newly determined the complete mitochondrial genome of N. yoldii, a marine gastropod mollusk in the family Neritidae. The specimen of N. yoldii was collected in Xiangshan, Zhejiang province, China (121.8°E, 29.5°N) and identified by morphology and deposited in Zhejiang Ocean University. The total DNA extraction was utilized the salting-out method (Aljanabi and Martinez Citation1997) with the muscle. We sequenced the mitochondrial genome on Illumina HiSeq X Ten platform using the total genomic DNA, and the genomic DNA was prepared in 400 bp paired-end libraries.
The annotated mitogenome of N. hepaticus has been deposited into GenBank database (GenBank accession number MK395169). The complete mitochondrial genome of N. yoldii is 15,791 bp in length. The complete mitochondrial genome has 13 protein-coding genes, 22 transfer RNA genes (tRNA), and two ribosomal RNA genes (12S rRNA and 16S rRNA). The overall base composition is A 29.5%, C 13.8%, G 21.5%, and T 35.2%. In 13 protein-coding genes, the nad2, nad5, and nad6 gene start with ATT, whereas nad4 started with ATA, the other nine genes start with ATG. For the stop codon, COX1, COX3, nad3, and atp8 end with TAG, the other nine genes end with TAA. The 16S rRNA is 1285 bp between the tRNALeu1 and tRNAVal, and the 12S rRNA is 869 bp between the tRNAVal and tRNAMet.
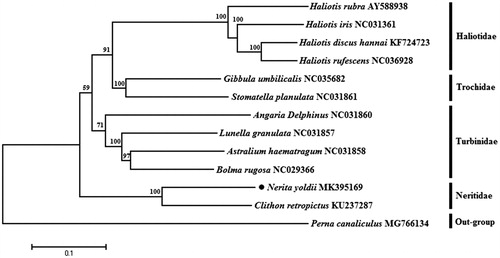
Phylogenetic analysis was performed using the sequences of 13 mitochondrial protein-coding genes and other 11 Archaeogastropoda species. Using Perna canaliculus of Mytilidae family as an outgroup (). A neighbour-joining (NJ) tree was performed with Mega 7.0 (Kumar et al. 2016). The tree showed that the N. yoldii is closely related to Clithon retropictus and belongs to the family Neritidae. We expect the present results will further facilitate for the study on the taxonomy, population genetic structure and phylogenetic relationships of Neritidae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693.
- Kumar S, Stecher G, Tamura KJ, 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. 33, 1870.
- Récluz C. 1841. Description de quelques nouvelles espèces de Nérites vivantes (Suite). Revue Zoologique. 4:147–152.
- Wang J, Yan H-Y, Cheng Z-Y, Huang X-W, Wang W, Ding M-W, Dong Y-W. 2018. Recent northward range extension of Nerita yoldii (Gastropoda: Neritidae) on artificial rocky shores in China. J Molluscan Stud. 84:345–353.
- Yeung CY, Yang SN. 2006. The ecology of Nerita yoldii and N. albicilla on Hong Kong rocky shores. PhD thesis, University of Hong Kong.
- Zhang X, QI CY, Zhang FS, Ma ST. 1963. A preliminary study of the demarcation of marine molluscan faunal regions of China and its adjacent waters. Oceanol Limnol Sinica. 2:124–138.