Abstract
The complete mitochondrial genome of Plestiodon tunganus from its type locality (Pengba town, Luding County, China) was determined using polymerase chain reaction and directly sequenced with a primer walking method. The mitogenome is 17,263 bp in length and comprises the standard set of one control region, two rRNAs, 22 tRNAs plus 13 protein-coding genes (PCGs). The PCGs were used to perform Bayesian phylogenetic analyses together with other scincid lizards with mitogenome data in GenBank. The resulting phylogenetic tree showed that P. tunganus clustered in a branch close to the species from the same genus. The mitogenome of P. tunganus will provide fundamental data to explore mitochondrial genome evolution in skinks.
Blue-tailed skinks (genus Plestiodon) are a common component of the terrestrial herpetofauna throughout their range in East Asia and North and Middle America (Brandley et al. Citation2005, Citation2011, Citation2012). There are 10 described species of Plestiodon in China (Cai et al. Citation2015, Kurita et al. Citation2017). Among them, P. tunganus is endemic to the western region in Sichuan Basin (Zhao and Adler Citation1993). So far, little is known about the mitochondrial genome evolution in Plestiodon lizards, even in scincid lizards.
In this study, we report the whole mitochondrial DNA genome of P. tunganus, with voucher number MCB1021. The specimen was collected from the type locality of Pengba town (30.02559°N, 102.18163°E), Luding County, Sichuan Province, China in August 2007. Its liver tissue was fixed with 95% ethanol and stored at –20°C in the herpetological collection at Chengdu Institute of Biology, Chinese Academy of Sciences. Total genomic DNA was extracted from liver tissue using a high-salt extraction protocol (Aljanabi and Martinez Citation1997). Published sequences of P. egregious (GenBank accession number AB016606), P. chinensis (Zhang et al. Citation2016) and P. elegans (Song et al. Citation2016) were used to design 16 pairs of primers. To obtain a sequence of the control region, we also designed some species-specific genome walking primers. Genes were annotated with the MITOS (Bernt et al. Citation2013) web server.
The mitochondrial genome of P. tunganus is 17,263 bp in length, with overall base composition of 24.5% (T), 29.5% (C), 30.4% (A) and 15.5% (G). Annotation of the mitogenome revealed 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one control region (CR or D-loop). The gene organization and order exhibited a typical vertebrate mitochondrial genome feature. Most genes are encoded on heavy strand except for ND6 and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser[UGA], Glu, and Pro). Most of the PCGs are initiated with the typical ATG codon, except for COX1 with GTT. Meanwhile, most PCGs are terminated with the typical AGA/AGG/TAA codons, except for COX2, COX3, ND3, and ND4 with the incomplete termination codon T. The 22 tRNA genes range in size from 66 bp in tRNA-Cys to 75 bp in tRNA-Phe and tRNA-Leu (UUA). The 12S rRNA, 16S rRNA, and D-loop are 950 bp, 1537 bp, and 2743 bp in size, respectively.
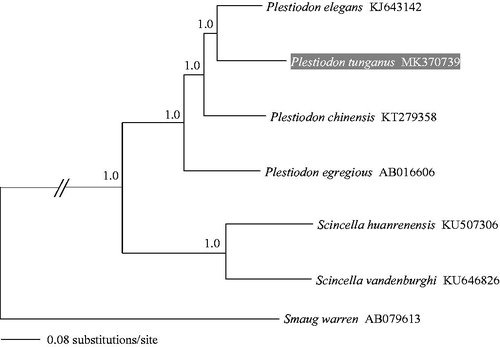
To evaluate mitochondrial sequence authenticity of P. tunganus and its phylogenetic placement, we performed a phylogenetic analysis using six complete mitochondrial genomes of the skinks (Squamata, Scincomorpha, Scincidae) as well as Smaug warreni (Squamata, Scincomorpha, Cordylidae) as the outgroup. We concatenated the PCGs and performed the Bayesian inference with MrBayes v.3.2.1 (Ronquist & Huelsenbeck Citation2003) with GTR + Gamma substitution model. The resulting phylogenetic tree () indicated a close phylogenetic affinity of the four Plestiodon species, confirming previous findings (Brandley et al. Citation2011, Citation2012). The mitogenome sequence of P. tunganus will provide fundamental data to explore mitochondrial genome evolution in skinks.
Nucleotide sequence accession number
The complete mitochondrial genome sequence of Plestiodon tunganus has been assigned GenBank accession number MK37039.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Figure 1. A majority-rule consensus tree inferred from Bayesian inference using MrBayes v.3.2.1 (Ronquist & Huelsenbeck Citation2003) based on the PCGs of four species of Plestiodon, two species of Scincella and one outgroup. The phylogeneic placement of P. tunganus is highlighted. DNA sequences were aligned in MEGA v.6.06 (Tamura et al. Citation2013). The PCGs were translated into amino acids sequences and were manually concatenated all sequences into a single nucleotide dataset (total 11,426 bp). Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution. GenBank accession numbers are given with species names.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25:4692–4693.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Brandley MC, Ota H, Hikida T, Nieto Montes de Oca A, Fería-Ortíz M, Guo X, Wang Y. 2012. The phylogenetic systematics of blue-tailed skinks (Plestiodon) and the family Scincidae. Zool J Linn Soc. 165:163–189.
- Brandley MC, Schmitz A, Reeder TW. 2005. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol. 54:373–390.
- Brandley MC, Wang Y, Guo X, Montes de Oca A, Fería-Ortíz M, Hikida T, Ota H. 2011. Accommodating heterogenous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Syst Biol. 60:3–15.
- Cai B, Wang YZ, Chen YY, Li JT. 2015. A revised taxonomy for Chinese reptiles. BIodivers Sci. 24:500–551.
- Kurita K, Hidetoshi O, Tsutomu H. 2017. A new species of Plestiodon (Squamata: Scincidae) from the Senkaku Group, Ryukyu Archipelago, Japan. Zootaxa. 4254:520–536.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Song T, Zhang C, Huang X, Zhang B. 2016. Complete mitochondrial genome of Eumeces elegans (Squamata: Scincidae). Mitochondrial DNA A. 27:719–720.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang C, Sun X, Chen L, Xiao W, Zhu X, Xia Y, Chen J, Wang H, Zhang B. 2016. The complete mitochondrial genome of Eumeces chinensis (Squamata: Scincidae) and implications for Scincidae taxonomy. Mitochondrial DNA A. 27:4691–4692.
- Zhao EM, Adler K. 1993. Herpetology of China. Oxford, OH: Society for the Study of Amphibians and Reptiles.
