Abstract
Camponotus concavus Kim & Kim, 1994 is a large carpenter ant endemic to the Korean peninsula. We have determined the mitochondrial genome of C. concavus. The circular mitogenome of C. concavus is 16,357 bp including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 985 bp. The base composition was AT-biased (77.7%). Gene order of C. concavus is identical to Camponotus atrox and differs in one tRNA from Polyrhachis dives, all of which belong to tribe Camponotini. Phylogenetic tree agrees with the current phylogenetic placement of C. concavus sister to all other Camponotus mitogenomes within the tribe Camponotini. The mitochondrial genome of C. concavus will be a good resource for understanding the species in aspect of molecular phylogeny.
Camponotus concavus, an endemic species of Korea, is a large carpenter ant commonly found in midland of the Korean peninsula. It usually nests in dead branches or logs in damp forests, but sometimes in rotting parts of living trees and rarely even in large cracks on rocks. They are mainly nocturnal foragers hunting for honeydew and insects, but few are also seen in daylight, especially when expanding their nest. Deeply concave clypeus and relatively sparse gastral pubescence making the abdomen slightly lustrous are unique characteristics to this species (Kim and Kim Citation1994). Nuptial flights occur around April to May, making it one of the earliest flying carpenter ants in Korea. Even though it is one of the dominant ant species of Korea, there is no intensive study for this species.
To understand genomic characteristics of C. concavus, we completed its mitochondrial genome from DNA extracted from a single virgin queen isolated at Sanghyo-dong, Seogwipo-si, Jeju-do, Republic of Korea using DNeasy Brood &Tissue Kit (QIAGEN, Hilden, Germany). Raw sequences obtained from HiSeq4000 (Macrogen Inc., Seoul, Korea) were filtered by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled by Velvet 1.2.10 (Zerbino and Birney Citation2008). Gaps were filled by SOAPGapCloser 1.12 (Zhao et al. Citation2011) and assembled sequences were confirmed with BWA 0.7.17 and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate the mitochondrial genome based on Camponotus atrox (NC_029357). ARWEN (Laslett and Canbäck Citation2008) was used to annotate tRNAs. The DNA sample and specimen (95% ethanol) were deposited in InfoBoss Cyber Herbarium (IN; Seoul, Republic of Korea; J. Park, KFDS00050).
Camponotus concavus mitochondrial genome length (Genbank accession is MK225553) is 16,357 bp. The nucleotide composition is AT-biased (A + T 77.7%). Its mitochondrial genome contains 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. The tRNAs’ size ranges from 63 to 94 bp, similar to other ants (circa 54-90 bp; Lee et al. Citation2018). Gene order of C. concavus is identical to those of Camponotus atrox (Kim et al. Citation2016) and only differs in the placement of tRNA-Ile from Polyrhachis dives (Liu et al. Citation2017) in tribe Camponotini. The control region, presumably corresponding to the single largest non-coding AT-rich region (985 bp, A + T 84.3%), is the third largest; control regions of C. atrox and Wasmannia auropunctata are 1402 bp and 987 bp, respectively (Kim et al. Citation2016; Duan et al. Citation2016).
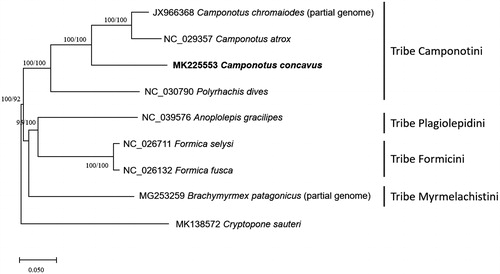
We inferred the phylogenetic relationship of eight Formicine ants including C. concavus and one outgroup species (Cryptopone sauteri; Park et al. Citation2019) using complete mitochondrial genome sequences. Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013). Bootstrapped neighbor-joining and maximum likelihood trees were constructed using MEGA X (Kumar et al. Citation2018). Phylogenetic position of C. concavus clustered with two Camponotus species (); however, tree topology inferred from complete mitochondrial genomes disagree with taxonomical classification at the level of tribal phylogeny (; Ward et al. Citation2016). Camponotus concavus mitochondrial genome will be a cornerstone for understanding the phylogenetic relationship of Camponotus species.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of all available ants mitochondrial genomes in Formicinae subfamily: Camponotus concavus (This study; MK225553), Camponotus atrox (NC_029357), Camponotus chromaiodes (JX966368; partial genome), Polyrhachis dives (NC_030790), Anoplolepis gracilipes (NC_039576), Formica fusca (NC_026132), Formica selysi (NC_026711), Brachymyrmex patagonicus (MG253259; partial genome), and Cryptopone sauteri (MK138572) as an outgroup species. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood methods.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Duan X-Y, Peng X-Y, Qian Z-Q. 2016. The complete mitochondrial genomes of two globally invasive ants, the Argentine ant Linepithema humile and the little fire ant Wasmannia auropunctata. Conserv Genet Resour. 8:275–277.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim B, Kim K. 1994. On the two new species, Camponotus concavus n. sp and fuscus n. sp from Korea (Hym., Formicidae). Korean J Entomol. 24:285–292.
- Kim MJ, Hong EJ, Kim I. 2016. Complete mitochondrial genome of Camponotus atrox (Hymenoptera: Formicidae): a new tRNA arrangement in Hymenoptera. Genome. 59:59–74.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lee C-C, Wang J, Matsuura K, Yang C-C. 2018. The complete mitochondrial genome of yellow crazy ant, Anoplolepis gracilipes (Hymenoptera: Formicidae). Mitochondrial DNA B. 3:622–623.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Liu J-H, Jia P-F, Fu J-Q, Dan W-L, Yang L-Y, Wang Q-M, Li Z-N. 2017. Characterization of mitochondrial genome and phylogenetic implications for Chinese black ant, Polyrhachis dives (Hymenoptera: Formicidae). Mitochondrial DNA B. 2:679–680.
- Park J, Kwon W, Park J. 2019. The complete mitochondrial genome of Cryptopone sauteri Wheeler, W.M., 1906 (Hymenoptera: Formicidae). Mitochondrial DNA B. 4:614–615.
- Ward PS, Blaimer BB, Fisher BL. 2016. A revised phylogenetic classification of the ant subfamily Formicinae (Hymenoptera: Formicidae), with resurrection of the genera Colobopsis and Dinomyrmex. Zootaxa. 4072:343–357.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
