Abstract
Blastus auriculatus, a shrub sparsely distributed in Yunnan, China, occurs only in bamboo forests at an elevation of below 200 m. It is categorized as ‘Critically Endangered (CR)’ in China Biodiversity Red List. In addition, the phylogenetic position of Blastus within the Melastomataceae family is still unclear. This study generated the complete chloroplast genome sequence of B. auriculatus with aims to provide genetic resources for conservation genetics and to reslove the phylogenetic position of Blastus. The size of the chloroplast genome of B. auriculatus is 155,981 bp, including a large single-copy (LSC) region of 85,955 bp and a small single-copy (SSC) region of 16,434 bp, separated by a pair of identical inverted repeat regions (IRs) of 26,796 bp each. The chloroplast genome contains a total of 125 genes, including 80 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that B. auriculatus is closest to Barthea barthei among species in Melastomataceae with available chloroplast genome sequences.
Blastus auriculatus Y. C. Huang, belonging to the family Melastomataceae, is a shrub sparsely distributed in Yunnan, China. It occurs only in bamboo forests below an elevation of 200 m (Hien and Thanh Citation2017). As the habitat of B. auriculatus is highly specialized, it has experienced rapid population contraction in the past few decades due to the degradation and loss of habitats. So far, B. auriculatus can be found in only two locations within an area of less than 100 km2. Being a rare species, it is categorized as “Critically Endangered (CR)” in China Biodiversity Red List (http://www.mee.gov.cn/gkml/hbb/bgg/201309/t20130912_260061.htm). Thus, it is necessary to scientifically protect this species which is at the high risk of extinction. To date, no studies have been conducted to comprehensively investigate the phylogenetic position of this genus within Melastomataceae. Genomic information of B. auriculatus would be valuable for conservation genetics of this species and resolution of the phylogenetic position of this genus. Here, we sequenced and characterized the complete chloroplast genome sequence of B. auriculatus as a resource for future studies.
Mature leaves of an individual of B. auriculatus were sampled from Hekou, Yunnan, China. The specimen is maintained in the Museum of Biology, Sun Yat-sen University (SYSBM) under the identification number of Y. Liu 613. Whole-genome DNA was extracted using a Plant Genomic DNA Kit (Magen, Guangzhou, China) and an Illumina paired-end (PE) library with an insert size of 350 bp was constructed and then sequenced on an Illumina HiSeq X Ten platform (Ige, Guangzhou, China).
After quality filtering, approximately 5.97 Gb high-quality reads were used to assemble the chloroplast genome using NOVOPlasty (Dierckxsens et al. Citation2017) with the rbcL sequence of Melastoma candidum (GenBank accession GQ436728) as the seed sequence. The chloroplast genome sequence of B. auriculatus was annotated using the DOGMA (Wyman et al. Citation2004) and submitted to GenBank with the accession number of MK335944.
The complete chloroplast genome of B. auriculatus has a circular molecular structure of 155,981 bp in length with 37.03% of GC content. It has a large single copy (LSC) region of 85,955 bp and a small single copy (SSC) region of 16,434 bp, separated by a pair of identical inverted repeat regions (IRs) of 26,796 bp each. The chloroplast genome contains a total of 125 genes, including 80 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
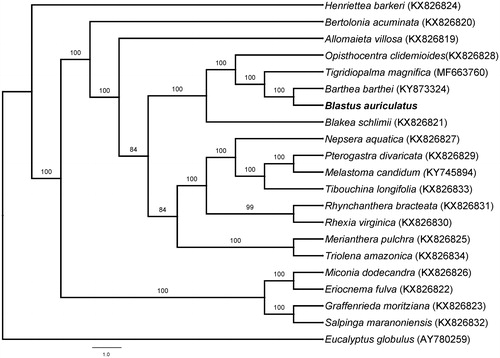
To clarify the phylogenetic position of Blastus within Melastomataceae, phylogenetic analyses were performed based on 20 species in Melastomataceae (Reginato et al. Citation2016; Ng et al. Citation2017; Zhou et al. Citation2018) as well as Eucalyptus globulus (Myrtaceae) as an outgroup. These sequences were aligned using MAFFT (Katoh and Standley Citation2013). The ML tree was produced by RAxML (Stamatakis Citation2014) using 1000 bootstrap replicates. As shown in the phylogenetic tree (), B. auriculatus is closest to Barthea barthei in the family Melastomataceae among closely related species with available chloroplast genome sequences. The chloroplast genome reported here offers a useful resource for conservation genetics and phylogenetic studies.
Figure 1. Maximum likelihood tree based on complete chloroplast genome sequences of 20 species of Melastomataceae with Eucalyptus globulus as an outgroup showing the phylogenetic position of Blastus auriculatus. The bootstrap support values shown next to the nodes were based on 1000 replicates. Scale in substitutions per site.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.8.
- Hien NT, Thanh NT. 2017. A taxonomic study of the genus Blastus Lour. (Melastomataceae) in Vietnam. VNU J Sci. 32:1S.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Ng WL, Cai Y, Wu W, Zhou R. 2017. The complete chloroplast genome sequence of Melastoma candidum (Melastomataceae). Mitochondr DNA B. 2:242–243.
- Reginato M, Neubig KM, Majure LC, Michelangeli FA. 2016. The first complete plastid genomes of Melastomataceae are highly structurally conserved. PeerJ. 4:e2715
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhou QJ, Ng WL, Wu W, Zhou RC, Liu Y. 2018. Characterization of the complete chloroplast genome sequence of Tigridiopalma magnifica (Melastomataceae). Conservation Genet Resour. 10:571–573.
