Abstract
The complete mitochondrial genome of Marmota himalayana was determined based on next-generation sequencing. Genome length consisted of 16,442 bp, with a base composition of 32.1% A, 31.4% T, 23.7% C, and 12.8% G. It consists of 13 protein-coding genes, 22 tRNAs, 2 ribosomal RNAs, and a control region. The gene order and organization were similar to most of the other vertebrates. The molecular data here we presented could play a useful role to study the evolutionary relationships and population genetics of H. marmot.
The Himalayan marmot Marmota himalayana, a large squirrel of the genus Marmota, is widely distributed at elevations of 1900–5000 m around the Himalayan regions of India, Nepal, Pakistan, and the Qinghai-Tibetan Plateau of China (Nikol’skii and Ulak Citation2006). It is regarded as an important species in the plateau ecosystem since it provides foods for predators such as snow leopard Pantheva uncial (Oli et al. Citation1993). Reports of M. himalayana were mainly focused on ecology, ethology, and taxonomy (Armitage Citation1975, Citation1999; Barash Citation1989; Arnold Citation1990; Nikol’skii and Ulak Citation2006). To facilitate the future researches of taxonomic resolution, population genetics, and phylogeography, the complete mitochondrial genome of M. himalayana was sequenced and characterized using NGS technologies (GenBank accession number MK305281).
Two specimens were collected from Longbao Town, Yushu Tibetan autonomous prefecture, Qinghai Province, China (33°29′80″N, 96°47′40″ E). Genomic DNA was extracted from muscle tissues using Ezup Column Animal Genomic DNA Kit (Sangon, Shanghai, China) following the protocol. The genomic DNA was stored at -80 °C in Medical College of Qinghai University. The whole genome shotgun method was used to construct the library and next-generation sequencing technique was used to sequencing based on the Illumina MiSeq sequencing platform (Illumina HiSeq 1500, Illumina, San Diego, CA, USA). Genes were annotated with DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013) and manually verified.
The complete mitochondrial genome of M. himalayana is 16,442 bp in length. The overall nucleotide composition is A (32.1), T (31.4%), C (23.7%), and G (12.8%), with a significant AT bias of 63.5%. It consists of 13 typical vertebrate protein-coding genes, 22 tRNA, 2 rRNA genes, and 1 control region. ND6 gene and 8 tRNA genes are encoded on the light (L) strain; the remaining genes are encoded on the heavy (H) strain. The typical ATN (ATG or ATT or ATA) start codons are present in PCGs. TAA and AGA stop codons are used for most genes, with the exception of incomplete stop codon T for COX3, ND2, and ND4, and TA for ATPase6, ND1, and ND3. The two rRNA genes are 971 bp and 1567 bp, respectively. All tRNAs were recognized by tRNAscan-SE1.21 (Lowe and Eddy Citation1997). They have the typical cloverleaf secondary structures except for the shortest tRNASer (AGN) which lacks the DHU arm. No tandem repeat is found in 924 bp-long control region.
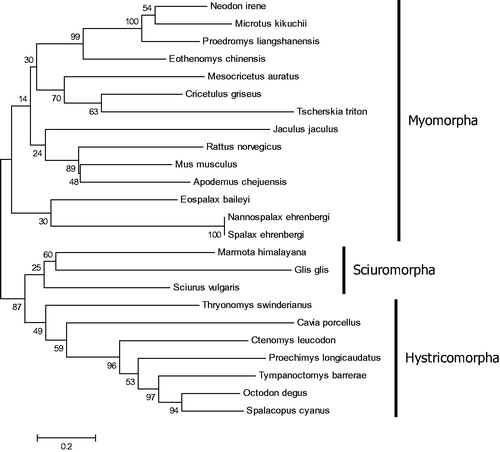
To understand the phylogenetic position of M. himalayana, phylogenetic analysis was performed by MEGA 6.06 (Tamura et al. Citation2013) based on the cytochrome c oxidase subunit I (COI) sequence of M. himalayana and those of 23 closely related species using maximum likelihood (ML) method with 1000 bootstrap replicates (). Different species from the same suborder clustered together (e.g. Myomorpha, Sciuromorpha, and Hystricomorpha). M. himalayana clustered within the suborder Sciuromorpha.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arnold W. 1990. The evolution of marmot sociality: II. Costs and benefits of joint hibernation. Behav Ecol Sociobiol. 27:239–246.
- Armitage KB. 1975. Social behavior and population dynamics of marmots. Oikos. 26:341–354.
- Armitage KB. 1999. Evolution of sociality in marmots. J Mammal. 80:1–10.
- Barash DP. 1989. Marmots: social behavior and ecology. Stanford Calif: Stanford University Press.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Nikol’skii AA, Ulak A. 2006. Key factors determining the ecological niche of the Himalayan marmot, Marmota himalayana Hodgson (1841). Russ J Ecol. 37:46–52.
- Oli MK, Taylor IR, Rogers ME. 1993. Diet of the snow leopard (Panthera uncia) in the Annapurna Conservation Area, Nepal. J Zool London. 231:365–370.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.