Abstract
Uca lactea is a crab belonging to the order Brachyura, but there is no report about its complete mitochondrial genome (mitogenome) in the literature. In this paper, the complete mitogenome of U. lactea was sequenced and the phylogenetic relationship was analyzed based on nucleotide sequences of the 13 protein-coding genes (PCGs) using the maximum likelihood (ML) method. The complete mitogenome of U. lactea is 15,659 bp (GenBank accession No. KY865330) in length and contains 22 tRNA genes, 13 protein-coding genes (PCGs), 2 rRNA genes, and a D-loop region. All PCGs were initiated by ATN codons. The nucleotide composition was also biased toward A + T nucleotides (69.48%). Phylogenetic analysis showed that U. lactea can be placed within the family Ocypodidae. This mitogenome sequence will provide a better understanding for crab evolution in the future.
Among extant crustaceans, Brachyuran crabs are one of the most species-rich clades with over 7000 described species (Guinot et al. Citation2013). The Uca lactea is mainly distributed in Korea, Japan, India, as well as Chinese Hainan, Fujian and other places. This is a species known as mostly living in the intertidal areas (Nakasone and Murai Citation1998). They are characterized by a strong sexual dimorphism and male asymmetry, males having a strongly enlarged cheliped used for fighting and signaling during courtship (Goshima and Murai Citation1988; Kim et al. Citation2004). Fiddler crabs walk sideways during feeding, slowly wander away from the burrow in a straight line (Zeil Citation1998). Mitogenome is a molecular marker used to find the evolution and phylogenetic relationships. So far, mitogenome of U. lactea has not been reported in the literature. For enriching the mitogenome library and providing the evidence of U. lactea taxonomic position, it was sequenced totally in our study.
In this report, we sequenced the complete mitogenome of U. lactea and reconstructed the phylogenetic relationship among other organisms through comparative genomics. U. lactea was collected from Fujian, China, and stored in our laboratory. The whole DNA was extracted using Aidlab Genomic DNA Extraction Kit (Aidlab, China) from the muscle. To amplify the mitogenome of U. lactea, six conserved genes (cox1, cox3, 12S, 16S, nad4, cob) were sequenced using the universal primer of Brachyura, specific PCR primers for U. lactea were designed, PCR temperature system and other conditions for extension were referred from the published article (Tang et al. Citation2017; Xin et al. Citation2017). Alignment of U. lactea and other Brachyura species sequences was performed using MAFFT (Katoh et al. Citation2002). The phylogenetic relationship was reconstructed based on the nucleotide sequences of 13 PCGs performed by the maximum likelihood (ML) method using IQ-tree (Nguyen Citation2015). The TIM + F + I + G4 model was used as the best model of nucleotide phylogenetic analysis chosen by ModelFinder (Kalyaanamoorthy et al. Citation2017).
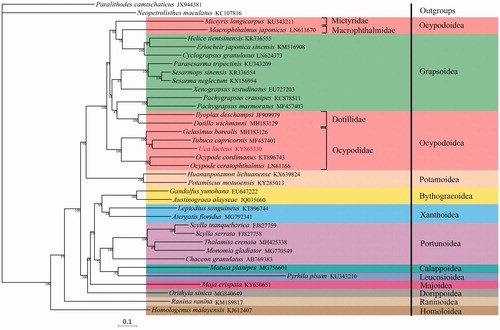
The data were submitted to the NCBI (KY865330). The size of the complete mitogenome of U. lactea was 15,659 bp, which included 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNA), 2 ribosomal RNAs (rRNA), and a D-loop region. There is an asymmetric nucleotide composition of U. lactea (A: 34.84%, T: 34.64%, G: 11.99%, C: 18.53%) that has a strong AT bias. Thirteen PCGs use diverse start codon, such as ATG (cox1, cox2, atp8, cox3, nad5, nad4L), ATT (atp6, nad6, nad1, nad2), ATC (nad3), GTG (nad4), ATA (cob). We reconstructed the phylogenetic tree based on nucleotide sequences of the 13 PCGs sequences using ML (). The result assuredly showed that U. lactea was close to Tubuca capricornis and Gelasimus borealis as a higher support, which belongs to Ocypodidae, Ocypodoidea of Brachyura. So, U. lactea appertain to Ocypodidae of Ocypodidae in Brachyura, and Ocypodidae is close to the superfamily of Grapsoidea. None of them is a monophyletic group observed from the phylogenetic tree, and it shows that Ocypodoidea and Grapsoidea are both reciprocal paraphyly consistent with previous research (Chen et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by authors.
Additional information
Funding
References
- Chen JQ, Xing YH, Yao WJ, Zhang C, Zhang Z, Jiang G, Ding Z. 2018. Characterization of four new mitogenomes from Ocypodoidea & Grapsoidea, and phylomitogenomic insights into thoracotreme evolution. Gene. 675:27–35.
- Goshima S, Murai M. 1988. Mating investment of male fiddler crabs, Uca lactea. Animal Behaviour. 36:1249–1251.
- Guinot DM, Tavares M, Castro P. 2013. Significance of the sexual openings and supplementary structures on the phylogeny of Brachyuran crabs (Crustacea, Decapoda, Brachyura), with new nomina for higher ranked podotreme taxa. Zootaxa. 3665:1–414.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 14:587–589.
- Katoh K, Misawa K, Kuma K-i, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kim TW, Christy JH, Choe JC. 2004. Semidome building as sexual signaling in the fiddler crab Uca Lactea (Brachyura: Ocypodidae). J Crustacean Biol. 24:673–679.
- Nakasone Y, Murai M. 1998. Mating behavior of Uca lactea perplexa (Decapoda: Ocypodidae). J Crustacean Biol. 18:70–77.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Tang B-P, Xin Z-Z, Liu Y, Zhang D-Z, Wang Z-F, Zhang H-B, Chai X-Y, Zhou C-L, Liu Q-N. 2017. The complete mitochondrial genome of Sesarmops sinensis reveals gene rearrangements and phylogenetic relationships in Brachyura. PLoS One. 12:e0179800.
- Xin ZZ, Liu Y, Zhang DZ. 2017. Mitochondrial genome of Helice tientsinensis (Brachyura: Grapsoidea: Varunidae): gene rearrangements and higher-level phylogeny of the Brachyura. Gene. 627:307–314.
- Zeil J. 1998. Homing in fiddler crabs (Uca lactea annulipes and Uca vomeris : Ocypodidae). J Compar Physiol A. 183:367–377.