Abstract
The complete mitochondrial genome of Channa gachua was determined based on next-generation sequencing. Genome length consisted of 16,561 bp, with a base composition of 28.3% A, 26.8% T, 29.3% C, and 15.6% G. It consists of 13 protein-coding genes, 22 tRNAs, 2 ribosomal RNAs, and a control region. The gene order and organization were similar to most of the other vertebrates. The molecular data here we presented could play a useful role to study the evolutionary relationships and population genetics of Channa gachua.
The dwarf snakehead Channa gachua is an important food fish and is one of the most suitable Channa species for aquarium due to its beautiful colouration and small size (Talwar and Jhingran Citation1992). This small air-breathing fish is commonly found in the shallow regions of streams and rivers, littoral of lakes and reservoirs, marshes, paddy fields, irrigation canals, and ditches. It usually feeds on aquatic insects and insect larvae, but also feeds on other small fish (Costa and Fernando Citation1967). Even though it is widely distributed throughout South, East, and Southeast Asia, it has declined drastically in India and Endangered in some Asian countries like Singapore (Lim and Ng Citation1990; Milton et al. Citation2018). To facilitate the future researches of taxonomic resolution, population genetics, and resources conservation, the complete mitochondrial genome of C. gachua was sequenced and characterized in the present study (GenBank accession number MK371068).
The samples were collected from Guangzhou, China. After sampling, the specimens were stored in 90% ethanol and kept in Freshwater Fisheries Research Institute of Jiangsu Province. Thirty pairs of PCR primers were designed based on sequences from related fish C. argus (GenBank number GU937112) and C. maculata (GenBank number JX978724). Mitogenome database was annotated by DOGMA (Wyman et al. Citation2004) and MitoFish (Iwasaki et al. Citation2013).
The complete mitochondrial genome of C. gachua is 16,561 bp in length. The overall nucleotide composition is 28.3% for A, 26.8 for T, 29.3 for C, and 15.6% for G, with a slight AT bias of 55.1% and a strong bias against G. It consists of 13 typical vertebrate protein-coding genes, 22 tRNA, 2 rRNA genes, and 1 control region. ND6 gene and 8 tRNA genes are encoded on the light (L) strain; the remaining genes are encoded on the heavy (H) strain. The typical ATN (ATG or ATA) start codons are present in PCGs except for COI with GTG, and they had three types of stop codons (TAA, TA–, and T–), and the incomplete stop codons were presumably completed by posttranscriptional polyadenylation (Ojala et al. Citation1981). The two rRNA genes are 946 bp and 1683 bp, respectively. All tRNA genes were recognized by tRNAscan-SE1.21 (Lowe and Eddy Citation1997). They have the typical cloverleaf secondary structures except for the shortest tRNASer (AGN) which lacks the DHU arm. No tandem repeat is found in 908 bp long control region.
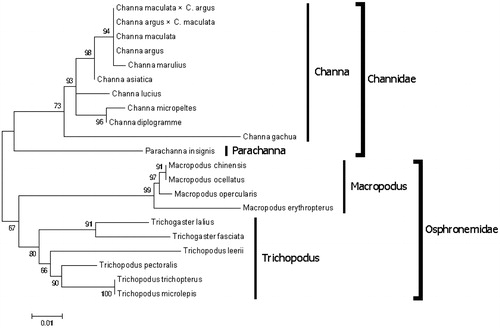
The cytochrome c oxidase subunit I (COI) sequence of C. gachua and other 20 closely related species were used for phylogenetic analysis by maximum likelihood methods with Mega 6.06 (Tamura et al. Citation2013). Different species from the same family clustered together (e.g. Channidae and Osphronemidae). C. gachua clustered with other snakehead fishes forming the genera Channa ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Costa HH, Fernando CM. 1967. The food and feeding relationships of the common meso and macrofauna in the Mahaoya, a small mountainous stream at Peradeniya, Ceylon. Ceylon J Sci. 7:7490.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Lim KKP, Ng P. 1990. The freshwater fishes of Singapore. Singapore: Singapore Science Centre; p. 160.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Milton J, Bhat AA, Haniffa MA, Hussain SA, Rather IA, Al-Anazi KM, Hailan WAQ, Abul Farah M. 2018. Ovarian development and histological observations of threatened dwarf snakehead fish, Channa gachua (Hamilton, 1822)). Saudi J Biol Sci. 25:149–153.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Talwar PK, Jhingran AG. 1992. Inland fishes of India and adjacent countries, vols. I and II. New Delhi, China: Oxford and IBH Publishing Company.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.