Abstract
The complete chloroplast genome of the Drooping Juniper, Juniperus recurva, was characterized. The cp genome was 127,602 bp in length, which contained 119 genes, including 82 protein-coding genes, 33 tRNA, and four ribosomal RNA genes. Eight genes (atpF, ndhA, ndhB, rpoC1, petD, petB, rpl16, and rpl2) had a single intron, whereas two genes (rps12 and ycf3) contained two introns. The GC content of this circle genome is 35.0%. Phylogenomic analysis based on 78 common protein sequences strongly supported the close relationship between J. recurva and J. tibetica.
Juniperus recurva Buch.-Ham. ex D. Don. is a large shrub or tree up to 40 m high that is native to the Himalaya, which distributed from northern Pakistan to western Yunnan in southwestern China at altitude from 2400 to 4500 m.a.s.l. (Farjon Citation2005). This species differs from its close relatives in its needle-like, six ranks leaves (in alternating whorls of three) and its pendulous and curved branchlets (Adams Citation2014). This species and its cultivars are widely cultivated as ornamental plants in Western Europe (Farjon Citation2005).
Fresh leaf materials were sampled from Langtang National Park, Nepal (28.1062° N, 85.3472° E) and immediately dried and kept in silica gel for DNA extraction. The dried leaf materials were deposited in the College of Life Sciences, Sichuan University (Sample No. Mao-NPL-63-01). Total genomic DNA was extracted using the modified CTAB method (Doyle and Doyle Citation1987) and sequenced by an Illumina Hiseq Platform (Illumina, San Diego, CA). In total, 17.38 M 150-bp raw paired reads were yielded. After removing the adapters, those reads were used to assemble the complete chloroplasts (cp) genome by NOVOPlasty (Dierckxsens et al. Citation2017) using the J. tibetica complete cp genome sequence as a reference. Plann v1.1 (Huang and Cronk Citation2015) and Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the cp genome and correct the annotation.
The cp genome sequence of J. recurva (GenBank accession no. MK375217) is 127,602 bp in length, containing 82 protein-coding genes, 33 tRNA genes, and four rRNA genes. Two genes (trnI-CAU and trnQ-UUG) occur in double copies, whereas the left 115 genes occur as a single copy. Among all of these genes, eight genes, atpF, ndhA, ndhB, rpoC1, petD, petB, rpl16, and rpl2, have a single intron, and two genes (rps12 and ycf3) contain two introns. Inverted repeat (IR) sequences were not detected in this juniper cp genome, which was consistent with previous analysis (Li et al. Citation2016; Tso et al. Citation2018). The overall GC content of the complete cp genome is 35.0%.
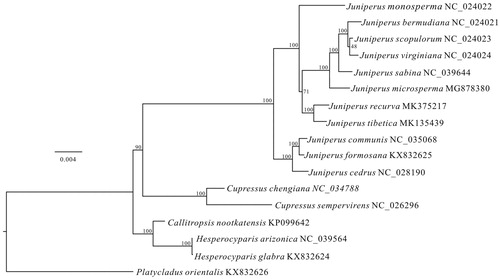
To further investigate the phylogenetic relationship between J. revurva and the other junipers, a maximum likelihood (ML) tree based on the concatenated amino acids of 78 protein-coding genes of 11 juniper species was constructed in RAxML v8 (Stamatakis 2014) with 200 bootstrap replicates. We selected six species of other four genera in Cupressaceae as outgroups and then aligned the sequences of all 17 speices via MAFFT v7 (Katoh and Standley Citation2013). Our results supported that J. revurva is closely related to J. tibetica (). The complete chloroplast genome of this ornamental juniper is useful to future evolutionary studies on conifers.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Adams RP. 2014. Junipers of the World: The genus Juniperus. 4th ed. Bloomington (IN): Trafford Publishing.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Farjon A. 2005. A monograph of Cupressaceae and Sciadopitys. Kew (United Kingdom): Royal Botanic Gardens.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li H, Guo Q, Zheng W. 2016. The complete chloroplast genome of Cupressus gigantea, an endemic conifer species to Qinghai-Tibetan Plateau. Mitochondrial DNA A. 27:3743–3744.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tso S, Li J, Xie S, Miao J, Hu Q, Mao K. 2018. Characterization of the complete chloroplast genome of Juniperus microsperma (Cupressaceae), a rare endemic from the Qinghai-Tibet Plateau. Conserv Genet Resour. https://doi.org/10.1007/s12686-018-1027-y.