Abstract
This paper reports the mitochondrial genome of Geokichla sibirica sibirica, which is a closed circular molecule of 16766 bp. All gene conformations were consistent with the gene arrangement in vertebrates. Phylogenetic analysis of cytochrome c oxidase subunit I and cytochrome b concatenated sequences from 10 species from seven genera was conducted using the neighbour-joining method and the Kimura 2-parameter model. The topology demonstrated that the mitogenome of this species was genetically closest to that of Zoothera aurea. Geokichla sibirica sibirica mitogenome can contribute to our understanding of the phylogeny and evolution of Zoothera and Geokichla.
Geokichla sibirica, which belongs to the genus Geokichla within the family Turdidae (Aves, Passeriformes), is mainly distributed in Northern Asia and it migrates through Southeast Asia to the Greater Sunda Islands during winter (Sangster, van den Berg, et al. Citation2009; Sangster, Collinson, et al. Citation2011). There are two subspecies included in G. sibirica – G. sibirica davisoni and G. sibirica sibirica. Geokichla sibirica sibirica breeds in Northeast China and Siberian coniferous forests, showing that it is highly migratory. Geokichla sibirica sibirica is similar in size to the song thrush. The species is omnivorous, eating a variety of insects, earthworms, and berries. Adult males and females differ in colour. Both males and females have a distinctive pattern of black stripes under their white wings, the same pattern seen in scaly thrushes (Bourski et al. Citation2015; Demidova Citation2011).
In this study, we determined the complete mtDNA sequence of G. sibirica sibirica. The samples were collected at the bird circle station of the Jiangsu Dafeng Milu National Natural Reserve in October 2018. After sampling, the specimens were stored in animal specimens museum of Nanjing Forestry University. Genome DNA was isolated from paw tissue using a kit and 12 primers were designed for fragment amplification.
The complete mitochondrial DNA of G. sibirica sibirica is a closed circular molecule composed of 16,766 bp (GenBank accession no. MK377247), containing the typical set of 22 transfer RNA genes, 13 protein-coding genes (PCGs), two ribosomal RNA genes (rrnS and rrnL), and one control region (D-loop). All the genes showed typical gene sequences consistent with vertebrates (Noack et al. Citation1996; Zeng et al. Citation2019). Among the 13 PCGs, in addition to ND2 having ATT and COX1 having ATG as their start codons, the other 11 PCGs had ATG as their start codon. Typical termination codons (TAA or TAG) appeared in 8 PCGs (ND2, COX2, ATP8, ATP6, ND3, ND4L, CYTB, and ND6), and the remaining 5 had incomplete termination codons. The D-loop was 992 bp in length. The nucleotide composition of G. sibirica sibirica was relatively equal (A + T, 52.3%; C + G, 47.7%).
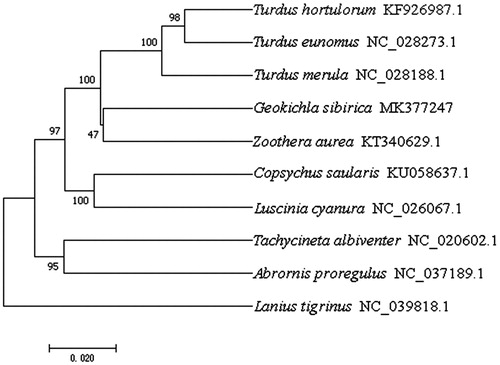
The complete mitochondrial genome sequence of the genus Geokichla has not been reported previously. In this study, phylogenetic analysis of the G. sibirica sibirica and nine other passeriformes birds was carried out based on the concatenated nucleotide sequences of cytochrome c oxidase subunit I and cytochrome b using the neighbour-joining method and the Kimura 2-parameter model in MEGA 7.0 with 1000 bootstrap replicates (Kumar et al. Citation2016). The phylogenetic tree indicated that the mitogenome of this species was genetically closest to that of Zoothera aurea (), which is in accordance with the current morphological classification. Geokichla sibirica sibirica mitogenome can contribute to our understanding of the phylogeny and evolution of Zoothera and Geokichla.
Disclosure statement
The authors report no conflicts of interest. The sequence has been submitted to NCBI under the accession no. MK377247.
Additional information
Funding
References
- Bourski OV, Demidova EY, Morkovin AA. 2015. Population dynamics of thrushes and seasonal resource partition. Biol Bull Rev. 5:220–232.
- Demidova EY. 2011. Reproductive parameters of four thrush species of Central Siberia. Biol Bull. 38:509–517.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Noack K, Zardoya R, Meyer A. 1996. The complete mitochondrial DNA sequence of the bichir (Polypterus ornatipinnis), a basal ray-finned fish: ancient establishment of the consensus vertebrate gene order. Genetics. 144:1165–1180.
- Sangster G, Collinson JM, Crochet PA, Knox AG, Parkin DT, Svensson L, Votier SC. 2011. Taxonomic recommendations for British birds: Fourth report. Ibis. 144:707–710.
- Sangster G, van den Berg AB, van Loon AJ, Roselaar CS. 2009. Dutch Avifaunal List: Taxonomic Changes in 2004–2008. Ardea. 97:373–381.
- Zeng L, Yang K, Wen A, Xie M, Yao Y, Xu H, Zhu G, Wang Q, Jiang Y, He T, et al. 2019. Complete mitochondrial genome of the Endangered fish Anabarilius liui yalongensis (Teleostei, Cyprinidae, Cultrinae). Mitochondrial DNA B. 4:21–22.