Abstract
The mitochondrial genome of Isaria tenuipes, strain TTZ2017-3, was sequenced on the Illumina Hiseq 4000 and the PacBio Sequel Sequencer and annotated. The genome is 66703 bp in length, encoding 15 conserved protein-coding genes (PCGs) including ribosomal protein S3, two rRNA genes and 26 tRNA genes. The nucleotide composition of I. tenuipes mitochondrial genome was 39.1% of A, 35.6% of T, 11.2% of C, 14.2% of G, 74.7% of A + T content. Phylogenetic analysis with other Hypocreales species revealed that I. tenuipes was more closely related to Cordyceps militaris, separated from Lecanicillium muscarium, Paecilomyces hepialid, and Beauveria species with Cordyceps teleomorph. This study provided valuable information on the gene contents of the mitochondrial genome and would facilitate the study of function and evolution of Isaria.
Isaria tenuipes (formerly Paecilomyces tenuipes) is an important entomogenous fungus which plays a dual role of biocontrol and medical value. As an insect pathogen, it usually has specificity for lepidopterous hosts (Fukatsu et al. Citation1997; Luangsa-Ard et al. Citation2005). Isaria tenuipes is also an anamorph of Cordyceps takaomontana Yakush & Kumaz (Spatafora and Blackwell Citation1993), which produce many bioactives with high medicinal value (Haritakun et al. Citation2007; Bunyapaiboonsri et al. Citation2011). The genus Isaria (Hypocreales: Cordypicitaceae) formerly belonged to Paecilomyces section Isarioidea and comprises a cosmopolitan group with high diversity of species and relatively broad host range, including arthropods and nematodes (Zimmermann Citation2008). Despite the great effort to use phylogenetic analyses based on multiple genes to identify Isaria species (Luangsa-Ard et al. Citation2005; Sung et al. Citation2007; D’Alessandro et al. Citation2014), there are still troubling gaps for such species complexes that urgently require sequence-based taxonomic clarification. Here, we report the complete mitochondrial DNA sequences of I. tenuipes (GenBank accession number: MH734936) for the first time, in order to provide valuable information on the gene contents of the mitochondrial genome for the study of function and evolution of Isaria.
Specimen of I. tenuipes was collected in Tiantangzhai Scenic Area (N31°11′49.1244″, E115°48′12.6288″), Lu’an Anhui, China, and isolated by Shengli Zhang. The isolated strain TTZ2017-3 is deposited in the Institute of Biological Engineering, Bozhou College, Anhui. Total mitochondrial genomic DNA was isolated by an improved extraction method (Chen et al. Citation2011) and sequenced on the Illumina Hiseq 4000 and the PacBio Sequel Sequencer and assembled using SPAdes v3.10.1 (Antipov et al. Citation2016). The assembled mitogenome of I. tenuipes was annotated as described previously (Zhang et al. Citation2017). Display quotations of over 40 words, or as needed.
The complete mitogenome sequence of I. tenuipes is 66703 bp in length, containing 39.1% of A, 35.6% of T, 11.2% of C, 14.2% of G, 74.7% of A + T content. There are 15 conserved protein-coding genes (PCGs), 26 tRNAs, two rRNA genes, and 14 ORFs, which were annotated. Those conserved PCGs includes rps3, ATP6 (1 group I intron), ATP8, ATP9, NAD1 (2 group I introns), NAD2 (2 group I introns), NAD3, NAD4 (1 group I introns), NAD4L, NAD5 (2 group IB introns), NAD6, COB (3 group I introns), COX1 (10 group IB introns), COX2 (2 group I intron), COX3 (2 group I introns). Two ribosomal RNA subunits (rns and rnl) contain 4 and 0 group I introns respectively. The 26 tRNAs covered 20 standard amino acids and clusterid into two clusters containing 23 tRNA genes. Three tRNA genes with different sequences and same anticodon (CAT) were found for tRNA-Met and two each were found for tRNA-Leu, tRNA-Arg, tRNA-Gly, and tRNA-Ser. For the remaining 15 tRNAs, only one gene each was found.
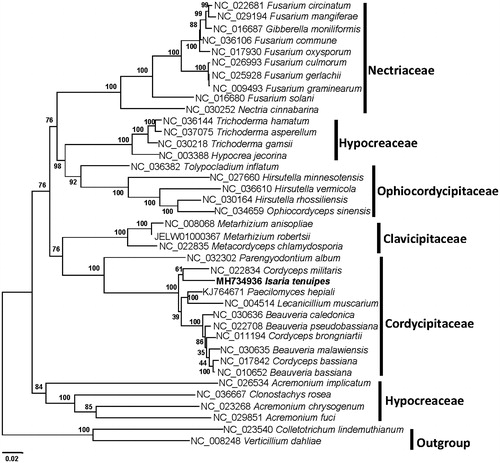
Phylogenetic analysis was conducted for the mitogenome and 36 other Hypocreales species using the Maximum Likelihood method of MEGA 7.0 with 1000 bootstrap replicates and GTR + I + G model (Kumar, Stecher, and Tamura Citation2016). The phylogenetic tree based on 14 PCGs indicated that the mitogenome of this species was genetically closest to that of Cordyceps molitaris (). It confirms that I. tenuipes is a member of Cordycipitaceae and supports Beauveria as an independent genus.
Figure 1. Phylogenetic relationships among 36 Hypocreales fungi inferred based on the concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The 14 mitochondrial protein-coding genes were nad1, nad2, nad3, nad4, nad4L, nad5, nad6, cox1, cox2, cox3, cob, atp6, atp8, and atp9. The tree was generated using Maximum Likelihood (ML). Numerical values along branches represent statistical support based on 1000 randomizations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Antipov D, Korobeynikov A, McLean JS, Pevzner PA. 2016. HYBRIDSPADES: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32:1009–1015.
- Bunyapaiboonsri T, Yoiprommarat S, Srisanoh U, Choowong W, Tasanathai K, Hywel-Jones NL, Luangsa-ard JJ, Isaka M. 2011. Isariotins G-J from cultures of the Lepidoptera pathogenic fungus Isaria tenuipes. Phytochem Lett. 4:283–286.
- Chen J, Guan R, Chang S, Du T, Zhang H, Xing H. 2011. Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. Plos One. 6:e17662.
- D’Alessandro CP, Jones LR, Humber RA, Lopez Lastra CC, Sosa-Gomez DR. 2014. Characterization and phylogeny of Isaria spp. strains (Ascomycota: Hypocreales) using ITS1-5.8S-ITS2 and elongation factor 1-alpha sequences. J Basic Microbiol. 54:S21–S31.
- Fukatsu T, Sato H, Kuriyama H. 1997. Isolation, inoculation to insect host, and molecular phylogeny of an entomogenous fungus Paecilomyces tenuipes. J Insect Pathol. 70:203–208.
- Haritakun R, Srikitikulchai P, Khoyaiklang P, Isaka M. 2007. Isariotins A-D, alkaloids from the insect pathogenic fungus Isaria tenuipes BCC 7831. J Nat Prod. 70:1478–1480.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Luangsa-Ard JJ, Hywel-Jones NL, Manoch L, Samson RA. 2005. On the relationships of Paecilomyces sect. Isarioidea species. Mycol Res. 109:581–589.
- Spatafora JW, Blackwell M. 1993. Molecular systematics of unitunicate perithecial ascomycetes: the clavicipitales-hypocreales connection. Mycologia. 85:912–922.
- Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol. 57:5–59.
- Zhang YJ, Yang XQ, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101:8517–8531.
- Zimmermann G. 2008. The entomopathogenic fungi Isaria farinosa (formerly Paecilomyces farinosus) and the Isaria fumosorosea species complex (formerly Paecilomyces fumosoroseus): biology, ecology and use in biological control. Biocontrol Sci Techn. 18:865–901.
