Abstract
The complete chloroplast (cp) genome sequence of Actinidia lanceolate has been characterized from Illumina pair-end sequencing. The complete cp genome was 156,390 bp in length, containing a large single copy region (LSC) of 90,032 bp, and a small single copy region (SSC) of 20,518 bp, which were separated by a pair of 22,920 bp inverted repeat regions (IR). The genome contained 131 genes, including 84 protein-coding genes, 39 tRNA genes, and eight rRNA genes. The phylogenetic position based on the chloroplast genome showed that A. lanceolate is sister to the group A. deliciosa and A. chinensis.
Actinidia lanceolate is a species of flowering plant in the family Kiwifruit. It is a small deciduous vine, densely rusted brown short hairs, visible in the pores, taupe in the next year, bald net hairless, small lenticels, and leaf paper. Its fruit tastes very good and is one of the best species of kiwifruit. For we know less about genomic information of A. lanceolate at present, getting the complete chloroplast genome is helpful to understand the evolutionary analysis of Kiwifruit. In this study, we assembled and characterized the complete chloroplast genome sequence of A. lanceolate based on the Illumina pair-end sequencing data and uncovered its phylogenetic relationship with eight Actinidiaceae species of which complete chloroplast genomes are available.
The voucher specimen of A. lanceolate was stored at Chinese Virtual Herbarium (CVH). Fresh leaves of ACH were collected from Shifang, Sichuan, China (31°13′45″ N, 104°0′55″ E). The total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987). The filtered reads were assembled by the program NOVOPlasty (Dierckxsens et al. Citation2017) for A. lanceolate using the sequenced plastome of its close relative Actinidia eriantha (GenBank accession no. KY100979) as the reference. The assembled plastome was annotated using Plann (Huang and Cronk Citation2015), and the annotation was corrected using Geneious (Kearse et al. Citation2012). The complete plastome together with gene annotations was submitted to GenBank under the accession number of MK370725.
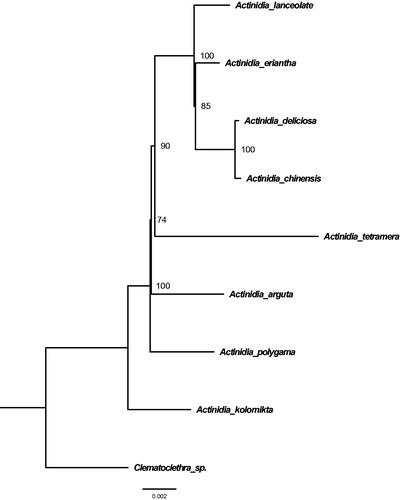
Our assembled plastome of A. lanceolate is 156,390 base pairs (bp) in size, containing a large single-copy (LSC) region of 90,032 bp, a small single-copy (SSC) region of 20,518 bp, and a pair of inverted repeat regions (IR) (IRa and IRb, 22,920 bp). In addition, the plastome possesses total 131 genes, including 84 protein-coding gene (PAZs, eight rRNA genes, and 39 tRNA genes). Most of these genes are in a single copy, however, one rRNA gene (i.e. 5S rRNA), three protein-coding genes (i.e. ndhB, rps12, and rps7), and eight tRNA genes (i.e. trnH-GUG, trnI-CAU, trnL-CAA, trnI-GAU, trnV-GAC, trnA-UGC, trnR-ACG, and trnN-GUU) occur in double copies. The overall GC-content of the whole plastome is 37.3% while the corresponding values of the LSC, SSC, and IR regions are 35.5, 31.1, and 43.4%, respectively. Furthermore, the phylogenetic position of A. lanceolate was determined using a Maximum Likelihood (ML) tree which was constructed based on whole chloroplast genome sequences of eight Actinidiaceae plants using RAxML (Kumar et al. Citation2016) with 1000 bootstrap replicates. All the sequences were first aligned using MAFFT (Katoh and Standley Citation2013). Our results showed that A. lanceolate is sister to the group A. deliciosa and A. chinensis ().
Figure 1. Phylogenetic relationships of nine seed plants based on plastome sequences. Bootstrap percentages are indicated for each branch. GenBank accession numbers: Actinidia arguta (NC_034913.1), Actinidia chinensis (NC_026690.1), Actinidia deliciosa (NC_026691.1), Actinidia eriantha (NC_034914.1), Actinidia kolomikta (NC_034915.1), Actinidia polygama (NC_031186.1), Actinidia tetramera (NC_031187.1), Actinidia lanceolate (Unpublished), and Clematoclethra scandens subsp. hemsleyi (KX345299.1).
The complete plastome can provide valuable information to develop highly variable DNA markers for population genetic survey, species delimitation, and other ecological and evolutionary studies. In addition, our study will benefit the study of the relationships between Kiwifruit species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
