Abstract
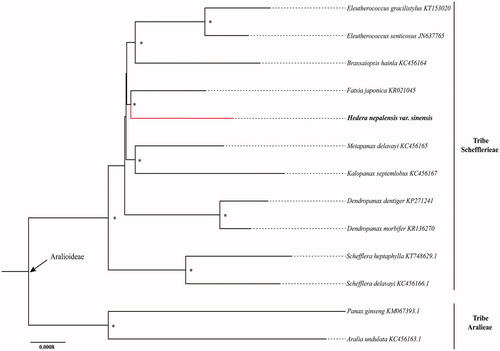
The complete chloroplast genome of widely cultivated Hedera nepalensis K. Koch var. sinensis (Tobl.) Rehder was sequenced and assembled in this study. The genome is 156,652 bp in length and contained 134 encoded genes in total, including 87 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. Based on available plastomes of 13 species in Araliaceae, we reconstructed their phylogenetic relationships and found that H. nepalensis var. sinensis is closely related to Fatsia japonica.
Hedera nepalensis K. Koch var. sinensis (Tobl.) Rehder, commonly known as Chinese ivy, is a perennially evergreen climbing shrub that belongs to the Araliaceae family. It is widely cultivated as an ornamental vine in eastern Asia, and has long been used as a herbal medicine for the treatment of diabetes (Saleem et al. Citation2014). Recent study reported it contains plenty of phenol chemical compound such as catechin and caffeic acid, which show significant antioxidant potential (Jafri et al. Citation2017). However, a few molecular and genetic resources are available for this heavily cultivated species. In this study, we assembled and characterized the complete chloroplast genome of H. nepalensis var. sinensis, which will provide organelle molecular basis for this valuable medicinal and horticultural plant.
The sample was collected from Xuancheng, Anhui, China (Voucher No. ZSTU01319, deposited at Zhejiang Sci-Tech University). DNA was isolated by the modified method CTAB assisted with DNA Plantzol (Invitrogen, Carlsbad, CA) from fresh leaves. The plastome sequences were generated using Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA). In total, ca. 20 million high-quality clean reads (150bp PE read length) were generated with adaptors trimmed. Then, NOVOPlasty (Dierckxsens et al. Citation2017) was used de novo to assemble the chloroplast genome with Fatsia japonica rbcL gene (Accession: U50245) as seed. Finally, we aligned and annotated the plastome with GENEIOUS v 9.0.6 (Biomatters Ltd, Auckland, New Zealand).
The full length of chloroplast genome H. nepalensis var. sinensis (GenBank accession No. MK130890) is 156,652 bp (with 38% GC content). It consists of a large single-copy region (LSC, 86597 bp, 36.2% GC content), a small single-copy region (SSC, 18,173 bp, 32% GC content), and two inverted repeat regions (IR, 25941 bp, 43.1% GC content). In total, there were 134 genes in H. nepalensis var. sinensis cp genome, including 87 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Among them, seven protein-coding genes (rpl2, rpl23, ycf2, ycf15, ndhB, rps7, rps12), all four rRNA genes (rrn16, rrn23, rrn4.5, rrn5), and seven tRNA genes (trnH-CAU, trnL-CAA, trnV-GAC, trnL-CAU, trnA-UGC, trnR-ACG, trnN-GUU) have two copies. Ten of these protein-coding genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, ndhA, rps12) have one intron each and two (ycf3, clpP) of them have two introns.
Thirteen species in Aralioideae with available chloroplast genomes were selected to study the phylogentic placement of H. nepalensis var. sinensis in Schefflerieae tribe (). The sequence alignment was conducted using MAFFT v7.3 (Katoh and Standley Citation2013). The phylogenetic tree was drawn using IQtree (Nguyen et al. Citation2015) with 5000 bootstrap replicates and K3Pu + F+R2 model. The phylogenetic tree revealed that H. nepalensis var. sinensis pertain to tribe Schefflerieae and is sister to Fatsia japonica.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Jafri L, Saleem S, Ihsan ul H, Ullah N, Mirza B. 2017. In vitro assessment of antioxidant potential and determination of polyphenolic compounds of Hedera nepalensis K. Koch Arab J Chem. 10:S3699–S3706.
- Katoh K, Standley DM. 2013. MAFFT (Multiple Sequence Alignment Software Version 7): improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Saleem S, Jafri L, ul Haq I, Chang LC, Calderwood D, Green BD, Mirza B. 2014. Plants Fagonia cretica L. and Hedera nepalensis K. Koch contain natural compounds with potent dipeptidyl peptidase-4 (DPP-4) inhibitory activity. J Ethnopharmacol. 156:26–32.