Abstract
Potentilla stolonifera var. quelpaertensis Nakai has developed stolon in comparison to Potentilla fragarioides var. major. To uncover phylogenetic relations of two species, we presented complete chloroplast genome of P. stolonifera var. quelpaertensis which is 156,393 bp long and has four subregions: 85,763 bp of large single-copy (LSC) and 18,590 bp of small single-copy (SSC) regions are separated by 26,020 bp of inverted repeat (IR) regions including 129 genes (84 protein-coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.9% and those in the LSC, SSC, and IR regions are 34.8%, 30.7%, and 42.7%, respectively. Phylogenetic trees show that P. stolonifera var. quelpaertensis is closely clustered with P. fragarioides var. major and Potentilla centigrana, of which morphological characteristics are clearly distinct with each other. Additional researches are strongly required to understand the relationship between morphological classification and phylogenetic relationship of these Potentilla species.
The genus Potentilla in tribe Potentilleae (Rosaceae) covers about 1,000 species mainly in the temperate region of the Northern Hemisphere and a few species in the Southern hemisphere (Eriksson et al. Citation1998; Soják Citation2004, Citation2008). Hybridization and allopolyploid events and apomixis are reported in Potentilla (Asker Citation1971; Soják Citation1985; Eriksen Citation1996), indicating complete chloroplast genome is important to uncover their phylogenic relationships.
Potentilla stolonifera var. quelpaertensis is a very small plant with highly developed stolon distributed in Jeju island (Nakai Citation1914). Potentilla stolonifera var. quelpaertensis and P. fragarioides var. major have only pinnately compound leaves but are distinguished by the presence of stolon. The latter is distributed inside Korean peninsula. Size of P. stolonifera var. quelpaertensis is various and are sometimes overlapped with size of P. fragarioides var. major (Heo Citation2009). Interestingly, phylogeny based on ITS and chloroplast markers presented that two species belong to the same lineage but are not monophyletic (Heo et al., under revision). To clarify the phylogenetic relationship of two species, chloroplast genome of P. stolonifera var. quelpaertensis was sequenced.
Total DNA of P. stolonifera var. quelpaertensis collected in Jeju island, Republic of Korea (Voucher in InfoBoss Cyber Herbarium (IN); K-I. Heo, IB-00575) was extracted from fresh leaves using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea, and de novo assembly and confirmations were done by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao Q-Y et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on P. centigrana chloroplast complete genome (MK209637; doi:10.1080/23802359.2019.1573113).
The chloroplast genome of P. stolonifera var. quelpaertensis (Genbank accession is MK229189) is 156,393 bp (GC ratio is 34.8%) and has four subregions: 85,763 bp of large single-copy (LSC; 34.8%) and 18,590 bp of small single-copy (SSC; 30.7%) regions are separated by 26,020 bp of inverted repeat (IR; 42.7%). It contained 129 genes (84 protein-coding genes, eight rRNAs, and 37 tRNAs); 17 genes (6 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
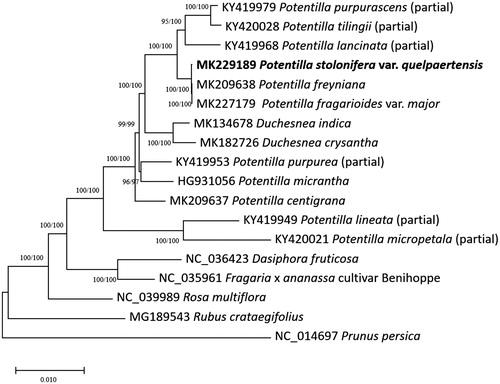
Eleven partial or complete Potentilla (Ferrarini et al. Citation2013; Zhang et al. Citation2017; https://www.tandfonline.com/doi/full/10.1080/23802359.2019.1573113; https://www.tandfonline.com/doi/full/10.1080/23802359.2019.1565977; Park et al. in submission; doi:10.1080/23802359.2019.1591202), two Duchesnea (https://www.tandfonline.com/doi/full/10.1080/23802359.2018.1553527; https://www.tandfonline.com/doi/full/10.1080/23802359.2019.1565964), and five Rosaceae chloroplast genomes (Cheng et al. Citation2017; Yang et al. Citation2017; Zhao et al. Citation2018) were used for constructing the neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees using MEGA X program (Kumar et al. Citation2018) based on sequence alignment of whole chloroplast genomes by MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees clearly show that P. stolonifera var. quelpaertensis and P. fragarioides var. major are clustered in one clade (). In addition, P. centigrana is morphologically distinguished with two species and is also clustered closely with two Potentilla species (), indicating additional researches to understand the relationship between morphological classification and phylogenetic relationship of three Potentilla species are strongly required.
Figure 1 Neighbor-joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 18 Rosaceae partial or complete chloroplast genomes: Potentilla stolonifera var. quelpaertensis (MK229189, in this study), Potentilla stolonifera (MK227179), Potentilla freyniana (MK209638), Potentilla centigrana (MK209637), Duchesnea chrysantha (MK182726), Duchesnea indica (MK134678), Potentilla tilingii (KY420028; partial genome), Potentilla lancinata (KY419968; partial genome), Potentilla purpurea (KY419953; partial genome), Potentila purpurascens (KY419979; partial genome), Potentilla micropetala (KY420021; partial genome), Potentilla micrantha (HG931056), Potentilla lineata (KY419949; partial genome), Dasiphora fruticosa (NC 036423), Fragaria × ananassa cultivar Benihoppe (NC_035961), Rubus crataegifolius (MG189543), Rosa multiflora (NC_039989), and Prunus persica (NC_014697). Phylogenetic tree was drawn based on neighbor joining phylogenetic tree. The numbers above the branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Asker S. 1978. Pseudogamy, hybridization and evolution in Potentilla. Hereditas. 87(2), 179–813.
- Cheng H, Li J, Zhang H, Cai B, Gao Z, Qiao Y, Mi L. 2017. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ. 5:e3919
- Eriksen B. 1996. Mating systems in two species of Potentilla from Alaska. Folia Geobot. 31:333–344.
- Eriksson T, Donoghue MJ, Hibbs MS. 1998. Phylogenetic analysis of Potentilla using DNA sequences of nuclear ribosomal internal transcribed spacers (ITS), and implications for the classification of Rosoideae (Rosaceae). Plant System Evol. 211:155–179.
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, Sargent DJ. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genom. 14:670.
- Heo K-I. 2009. A systematic study of genus Potentilla s.l. in Korea (Rosaceae). Suwon, Korea: Sungkyunkwan University.
- Heo et al. under revision. Taxonomic studies of the tribe Potentilleae (Rosaceae) in Korea.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Quant Biol. arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Nakai T. 1914. LIX. Plantae novae Coreanae et Japonicae. II. Fedde Repert Spec Nov Regni Veg. 13:275.
- Soják J. 1985. Notes on Potentilla: 1. Hybridogenous species derived from intersectional hybrids of sect. Niveae × Sect. Multifidae. Bot Jahrb Syst. 106:145–210.
- Soják J. 2004. Potentilla L.(Rosaceae) and related genera in the former USSR (identification key, checklist and figures) notes on Potentilla XVI. Bot Jahrb Syst. 125:253–340.
- Soják J. 2008. Notes on Potentilla XXI. A new division of the tribe Potentilleae (Rosaceae) and notes on generic delimitations. Bot Jahrb Syst. 127:349–358.
- Yang JY, Pak J-H, Kim S-C. 2017. The complete chloroplast genome sequence of Korean raspberry Rubus crataegifolius (Rosaceae). Mitochondrial DNA B. 2:793–794.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytologist. 214:1355–1367.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zhao Y, Lu D, Han R, Wang L, Qin P. 2018. The complete chloroplast genome sequence of the shrubby cinquefoil Dasiphora fruticosa (Rosales: Rosaceae). Conserv Genet Resour. 10:675–678.
