Abstract
Datong horse processes a muscular physique with high tolerance to extensive management and adapts well to high altitude conditions in Qinghai, China. In this study, the whole mitochondrial genome sequence of E. caballus determined using next-generation sequencing data was 16,654 bp in length, consisting of 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and a non-coding control region. Notably, there was one tandem repeat with 21 ‘CACCTGTG’ located in the control region. The base composition of the genome was A (32.3%), C (28.6%), G (13.3%), and T (25.8%), with an A + T content of 58.1%. Molecular phylogeny demonstrated that E. caballus was more closely related to a breed from central Asia and processed distinct relationships with some Chinese breeds. This work characterized the mitogenome of E. caballus (Datong horse breed) and present significant genetic information for this valuable specimen.
Datong horses (Equus caballus) are used for transportation, grazing, and riding and also involved in other activities such as tourism, competition, and entertainment in Qinghai, China. Datong horses currently show higher disease resistance, genetic stability, and adaptability to plateau conditions. Recently, a dramatic reduction of this species has been observed, due to urbanization and the popularization of mechanization. To enhance awareness regarding the protection of horse germplasm, it is urgent to assess Datong horse’s genetic diversity, evolution, and genetic relationships (Zeder Citation2008). Mitochondrial DNA (mtDNA) is an effective candidate maker due to characteristics of high mutation rates and matrilineal inheritance and has been widely applied to analyze origin and evolution, population genetic differentiation, and molecular phylogenetic properties (Jansen et al. Citation2002; Harrison and Turrion-Gomez Citation2006; Achilli et al. Citation2012; Olivieri et al. Citation2015). In this study, the complete mitochondrial genome sequence (GenBank: MG426188) of E. caballus was sequenced, assembled, and characterized, providing valuable molecular genetic evidence and scientific guidance for the protection and exploitation of E. caballus.
Specimen of E. caballus was obtained from Haiyan County, Qinghai province, China (N36°55′19ʺ, E100°56′44ʺ), and the blood sample was stored in the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS at −80 °C. Total DNA was isolated to prepare a genomic library; sequencing was performed on an Illumina HiSeq 2500 Sequencing System (Illumina, CA, USA) according to the standard operating protocol. Data assessment and quality filtering were conducted with CLC Genomics Workbench v10 (CLC Bio, Aarhus, Denmark). Then, high-quality clean reads were further used to assemble the complete mitochondrial genome with MITObim v1.9 (Hahn et al. Citation2013), using the previously reported Equus caballus mitogenome sequence (GenBank: KT781689) as an initial reference. A total of 1691 reads were mapped to the reference mitochondrial genome with a final coverage of 20.7x. The mitochondrial genome was annotated GENEIOUS R10 (Biomatters Ltd., Auckland, New Zealand). In brief, protein-coding genes (PCGs), tRNA genes, rRNA genes, and a control region (CR) were identified and annotated based on alignment, with counterparts from the mitochondrial genome of known E. caballus.
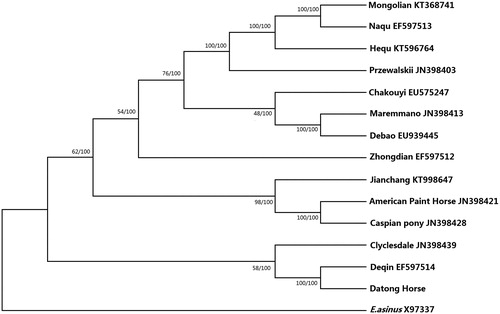
Sequencing of the entire mitochondrial genome showed a circular genome of 16,654 bp, containing 13 PCGs, two rRNA genes, 22 tRNA genes, and a non-coding control region, as well as the initiation site of origin L-strand replication (OL) comprising 32 bp between tRNA-Asn and tRNA-Cys. Meanwhile, there was a tandem repeat with 21 ‘CACCTGTG’ sequences located in the control region. The nucleotide contents of A, C, G, T, and A + T were 32.3%, 28.6%, 13.3%, 25.8%, and 58.1%, respectively. To determine the phylogenetic position of E. caballus Datong horse breed, phylogenetic analysis by two methods was conducted based on 14 mitochondrial genome from E. caballus and an available complete mitogenome of donkey as an outgroup; a similarity tree was generated based on the maximum likelihood () (Kavar and Dovc Citation2008; Tamura et al. Citation2011). Both NJ and ML trees showed identical topology and were well supported by bootstrap values. Molecular phylogenetic analyses suggested that Datong horse had a closer relationship with a breed from Central Asia and is distinctly related to some Chinese breeds, providing significant genetic information for exploring the evolution and history of E. caballus.
Figure 1. Phylogenetic analysis of selected Equidae species based on the maximum likelihood (ML) and neighbour-joining (NJ) methods. Concatenated coding sequences of 13 mitochondrial PCGs were assessed. The values at the nodes correspond to ML (right) and NJ (left) bootstrap support in percentages, respectively.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Achilli A, Olivieri A, Soares P, Lancioni H, Kashani BH, Perego UA, Nergadze SG, Carossa V, Santagostino M, Capomaccio S, et al. 2012. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc Natl Acad Sci USA. 109:2449–2454.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Harrison SP, Turrion-Gomez JL. 2006. Mitochondrial DNA: an important female contribution to thoroughbred racehorse performance. Mitochondrion. 6:53–63.
- Jansen T, Forster P, Levine MA, Oelke H, Hurles M, Renfrew C, Weber J, Olek K. 2002. Mitochondrial DNA and the origins of the domestic horse. Proc Natl Acad Sci USA. 99:10905–10910.
- Kavar T, Dovc P. 2008. Domestication of the horse: genetic relationships between domestic and wild horses. Livest Sci. 116:1–14.
- Olivieri A, Gandini F, Achilli A, Fichera A, Rizzi E, Bonfiglio S, Battaglia V, Brandini S, De Gaetano A, El-Beltagi A, et al. 2015. Mitogenomes from Egyptian cattle breeds: new clues on the origin of Haplogroup Q and the early spread of Bos taurus from the Near East. PLoS One. 10:e0141170.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis usingmaximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Zeder MA. 2008. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proc Natl Acad Sci USA. 105:11597–11604.
