Abstract
Cryptopygus antarcticus travei (Collembola) is a springtail endemic to the sub-Antarctic Prince Edward Islands. The mitogenome of C. a. travei has a length of 15,743 bp and comprises 13 protein-coding genes, 22 tRNAs, and two rRNAs. The base composition is 36% adenine, 33% thymine, 13% guanine, and 18% cytosine. Phylogenetic analyses confirmed the distinctness of C. a. travei from C. antarcticus, with considerable sequence divergences separating taxa within this group. In light of these results, we suggest that C. a. travei may be a candidate new species and that the current taxonomical status of this species should be re-evaluated.
Springtails (Class: Collembola) are a diverse, abundant, and widespread group. They occur in some of the harshest and most remote areas on earth, including Antarctica and the sub-Antarctic islands (Deharveng Citation1981; Hopkin Citation1997). The challenging conditions in these regions undoubtedly impose strong selection pressures (Holt and Gaines Citation1992; Manel et al. Citation2003), and biota continuously have to adapt to these ever-changing environments that are experiencing accelerated rates of climate change.
Not surprisingly, Collembola have been widely used to study biochemical and physiological adaptations (Sinclair et al. Citation2003). In the sub-Antarctic region, relatively few studies have documented genetic structures (Stevens et al. Citation2006; Myburgh et al. Citation2007), and only one attempted to link genetic diversity to physiological attributes (McGaughran et al. Citation2010). One of the most speciose genera in the region is Cryptopygus (Stevens and Hogg Citation2006; Carapelli et al. Citation2008). Cryptopygus antarcticus travei Deharveng, Citation1981 is restricted to the Prince Edward Islands where they reach densities of up to 104–105 individuals/m2 (Gabriel et al. Citation2001; Hugo et al. Citation2006). Here, we report the assembly and annotation of its mitogenome and evaluate its phylogenetic placement.
Total gDNA was extracted from 13 complete specimens sampled on the Prince Edward Islands (−46.767, 37.850) using the DNeasy® Blood and Tissue Kit (Qiagen®, Germany). Since whole specimens were used for DNA extractions, another specimen (collected from the same locality at the same time) serves as the voucher specimen (Iziko Museum, Cape Town, voucher number SAM-ENW-C010311). Genomic DNA libraries were prepared and indexed for each specimen. The resulting libraries were sequenced using the Illumina HiSeq4000 platform (Illumina, USA), producing 150 GB of raw data. The mitogenome was assembled from the pooled data using NOVOPlasty (Dierckxsens et al. Citation2017) with default settings, but with the insert size set to 350 bp. The cytochrome c oxidase subunit I (COI) gene of C. a. travei (Stevens et al. Citation2006; accession number: DQ285390) was used as the starting seed. The mitogenome was annotated using the MITOS2 WebServer (Donath et al. in press). The protein-coding genes (PCGs) and the ribosomal RNA boundaries were determined manually using Exonerate v2.2 (Slater and Birney Citation2005) and Infernal v1.1 (Nawrocki and Eddy Citation2013). Our reference data included the annotated mitogenomes of nine other species (>69% identity based on BLAST results).
The circularised mitogenome (15,743 bp in length) comprises 13 PCGs, 22 tRNAs, and two rRNAs. The base composition is 36% adenine, 33% thymine, 13% guanine, and 18% cytosine. An A + T rich region (69%) is present which corresponds to sites for replication initiation and transcription (Goddard and Wolstenholme Citation1978; Carapelli et al. Citation2008). Several instances of non-canonical start and incomplete stop codons (TA– or T–) were observed which is consistent with findings by Donath et al. (in press) and Carapelli et al. (Citation2008).
The 13 PCGs were aligned to data available for nine other species including five springtails and four hexapods. Bayesian phylogenetic analyses were performed in BEAST v2.5.0 (Bouckaert et al. Citation2014) using the HKY substitution model (Hasegawa et al. Citation1985) with a chain length of 300 million generations and a 33% burn-in. Resultant topologies were visualised in Figtree v1.4.3 (Rambaut Citation2016) ().
Figure 1. Bayesian phylogenetic tree of Cryptopygus antarcticus travei and nine other species (>69 identity) (five springtail species and four hexapod species). The following species and GenBank accession numbers used for the phylogenetic analysis are as follows: Cryptopygus antarcticus travei (MK433191), Cryptopygus antarcticus (NC_010533), Cryptopygus terranovus (NC_037610), Folsomia candida (KU198392), Folsomotoma octooculata (NC_024155), Gomphiocephalus hodgsoni (NC_005438), Anopheles arthuri (NC_037806), Delia antiqua (NC_028226), Simosyrphus grandicornis (NC_008754), and Elaphrus cupreus (KX087286). All the nodes on the tree are supported by a posterior probability of 1.
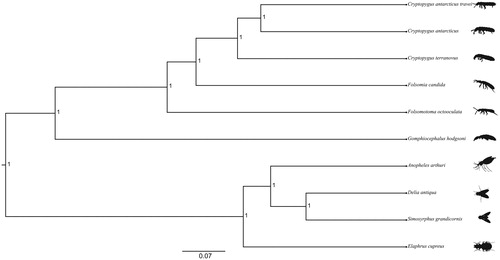
Collembola form a well-supported monophyletic clade, with C. a. travei as the sister taxon to C. antarcticus but with considerable genetic divergence separating them. Unusually high levels of sequence divergence for the COI gene was similarly reported by Stevens et al. (Citation2006) for Cryptopygus across the sub-Antarctic region. When taken together, these results strongly indicate that C. a. travei, confined to the Prince Edward Islands, is a candidate new species.
Acknowledgements
Samples were collected under permit from the Department of Environmental Affairs: (South African National Antarctic Programme) who also provided logistic support during the voyages. Analyses platforms and bioinformatics support was provided by the Centre for High Performance Computing (CHPC) in Cape Townand the University of Johannesburg IT services.
Disclosure statement
No conflict of interest is reported by the authors who are responsible for the content and writing of this paper.
Additional information
Funding
References
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10:e1003537.
- Carapelli A, Comandi S, Convey P, Nardi F, Frati F. 2008. The complete mitochondrial genome of the Antarctic springtail Cryptopygus antarcticus (Hexapoda: Collembola). BMC Genomics. 9:315–327.
- Deharveng L. 1981. Collemboles des Iles Subantarctiques de l’Ocean Indien. Comite Nationale Francais Des Recherches Antarctiques. 48:33–108.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:1–9.
- Donath A, Juhling F, Al-Arab M, Stadler PF, Middendorf M, Bernt M. In press. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 1–10.
- Gabriel AGA, Chown SL, Barendse J, Marshall DJ, Mercer RD, Pugh PJA, Smith VR. 2001. Biological invasions of Southern Ocean islands: the Collembola of Marion Island as a test of generalities. Ecography. 24:421–430.
- Goddard JM, Wolstenholme DR. 1978. Origin and direction of replication in mitochondrial DNA molecules from Drosophila melanogaster. Proc Natl Acad Sci. 75:3886–3890.
- Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 22:160–174.
- Holt RD, Gaines MS. 1992. Analysis of adaptation in heterogeneous landscapes: implications for the evolution of fundamental niches. Evol Ecol. 6:433–447.
- Hopkin SP. 1997. Biology of the Springtails (Insecta: Collembola). New York: Oxford University Press.
- Hugo EA, Chown SL, McGeoch MA. 2006. The microarthropods of sub-Antarctic Prince Edward Island: a quantitative assessment. Polar Biol. 30:109–119.
- Manel S, Schwartz MK, Luikart G, Taberlet P. 2003. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 18:189–197.
- McGaughran A, Convey P, Redding GP, Stevens MI. 2010. Temporal and spatial metabolic rate variation in the Antarctic springtail Gomphiocephalus hodgsoni. J Insect Physiol. 56:57–64.
- Myburgh M, Chown SL, Daniels SR, Jansen van Vuuren B. 2007. Population structure, propagule pressure, and conservation biogeography in the sub-Antarctic: lessons from indigenous and invasive springtails: biodiversity research. Divers Distrib. 13:143–154.
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 29:2933–2935.
- Rambaut A. 2016. Figtree 1.4.3. [accessed 2019 Jan 5] Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Sinclair BJ, Vernon P, Klok CJ, Chown SL. 2003. Insects at low temperatures: an ecological perspective. Trends Ecol Evol. 18:257–262.
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinform. 6:31–42.
- Stevens MI, Greenslade P, Hogg ID, Sunnucks P. 2006. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Southern hemisphere springtails: could any have survived glaciation of Antarctica? Mol Biol Evol. 23:874–882.
- Stevens MI, Hogg ID. 2006. The molecular ecology of Antarctic terrestrial and limnetic invertebrates and microbes. In: Huiskes A, Convey P, Bergstrom D, editors. Trends in Antarctic terrestrial and Limnetic ecosystems. Dordrecht (The Netherlands): Springer; p. 1–369.
