Abstract
Sinocyclocheilus yishanensis belongs to the genus Sinocyclucheilus (Cypriniformes, Cyprinidae), endemic to Guangxi province, Southwestern China. The circular mitogenome was 16,573 bp in length and consisted of 13 protein-coding genes, two ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and one non-coding control region. The overall base composition was 31.83% A, 25.14% T, 27.03% C, and 16.00% G with 43.03% GC content. Phylogenetic analysis using mitochondrial genomes of 16 species showed that all Sinocyclocheilus species clustered as one monophyletic clade except S. jii, and S. yishanensis was the closest to S. ronganensis.
Sinocyclocheilus yishanensis, a freshwater cyprinid fish belonging to the genus Sinocyclucheilus (Cypriniformes, Cyprinidae), was first found in the Lidong Reservoir (belong to Hongshui River) of Yishan county, Southwestern China (Li and Lan Citation1992). To date, there are 14 Sinocyclucheilus species of the complete mitogenome sequence which are available in Genbank, including S. yishanensis in this study. It will provide useful molecular data for genetics and evolutionary studies on this species and its closely related species.
The sample of S. yishanensis was collected from the underground river (belong to Liujiang River) located in Liujiang county of Guangxi, China (24°4′9.58′N, 108°59′14.05′E), and its muscle samples were fixed in 95% ethanol and preserved in Liuzhou Aquaculture Technology Extending Station, Liuzhou, China. Total genomic DNA was extracted from the fins through using traditional phenol-chloroform extraction method (Taggart et al. Citation1992). Then, we have sequenced the complete mitochondrial genome of S. yishanensis using the Illumina Hiseq4000 platform with de novo strategy (Tang et al. Citation2015) and submitted the genome into GenBank with accession number MK387704.
The circular mitogenome of S. yishanensis was 16,573 bp in length, including 13 protein-coding (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). The overall base composition was 31.83% A, 25.14% T, 27.03% C, and 16.00% G with 43.03% GC content. As previously reported, the Sinocyclocheilus species had the similar gene arrangement and nucleotide composition of the mitogenome (Li et al. Citation2018); besides, the locations of 37 genes on H-strand or L-strand and the start codon usage for 13 PCGs were consistent, including S. yishanensis in this study. The difference was the stop codon usage, 13 PCGs of the mitogenome of S. yishanensis had four types (TAA, TAG, TA-, and T–), and the incomplete stop codon (T–) appeared in those four PCGs (COII, ND3, ND4, and Cyt b).
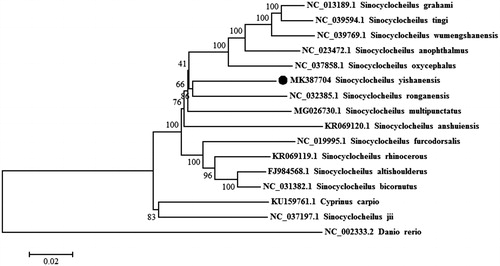
To validate the phylogenetic position of S. yishanensis, a neighbour-joining (NJ) tree inferred from the whole mitogenome of 14 Sinocyclocheilus species and two outgroup species (Cyprinus cario and Danio rerio) was constructed with 1000 bootstrap replicates using MEGA 6.06. The phylogenetic results revealed that all Sinocyclocheilus species clustered as one monophyletic clade except S. jii, and S. yishanensis was the most close to S. ronganensis (Luo et al. Citation2017), it was consistent with the BLAST results (92% identities to the mitogenome of S. ronganensis) and morphological comparison results ().
References
- Li CQ, Huang HT, Yang SF, He H, Fu QY, Chen SY, Xiao H. 2018. Complete mitochondrial genome and phylogenetic analysis of Sinocyclocheilus oxycephalus (Cypriniformes: Cyprinidae). Mitochondrial DNA B. 3:243–244.
- Li WX, Lan JH. 1992. A new genus and three new species of Cyprinidae from Guangxi, China. J Zhanjiang Fisheries Coll. 12:46–50.
- Luo FG, Huang J, He AY, Luo T, Zhou HJ, Wen YH. 2017. Complete mitochondrial genome of a cavefish Sinocyclocheilus ronganensis (Cypriniformes: Cyprinidae). Mitochondrial DNA B. 2:117–118.
- Taggart J, Hynes R, Prod Uhl P, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965.
- Tang M, Hardman CJ, Ji Y, Meng G, Liu S, Tan M, Yang S, Moss ED, Wang J, Yang C, et al. 2015. High-throughput monitoring of wild bee diversity and abundance via mitogenomics. Methods Ecol Evol. 6:1034–1043.