Abstract
The Yarkand toad-headed agama, Phrynocephalus axillaris, is endemic to an arid region in Northwest China from the Taklamakan Desert eastward to Dunhuang and Turpan depressions. A nearly complete mitochondrial genome of one individual from the Turpan Depression was determined using next-generation sequencing. The mitogenome is 15,883 bp in length, comprising two ribosomal RNA genes, 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), and a control region (CR). The gene arrangement and composition is similar to those of other Agaminae lizards, albeit with one part of CR (651 bp) existing between the tRNA-Thr gene and tRNA-Pro gene and another part of CR (with 107 bp already sequenced) inserting between the tRNA-Phe and 12S rRNA. Bayesian phylogenetic analyses confirmed the monophyly of both genus Phrynocephalus and its viviparous species. The mitogenome sequence may provide fundamental data to investigate altitude-related evolution of mitochondrial genes in Phrynocephalus.
The Yarkand toad-headed agama, Phrynocephalus axillaris Blanford, 1875, is endemic to an arid region in Northwest China from the Taklamakan Desert eastward to Dunhuang and Turpan depressions (Zhao et al. Citation1999). Recent advances in next-generation sequencing (NGS) technologies have facilitated the obtainment of mitochondrial genome sequences (Hahn et al. Citation2013). In this study, we determined a nearly complete mitochondrial genome of P. axillaris using NGS reads through Roche 454 sequencing platform. The lizard was collected from Turpan Desert Botanical Garden (N42.85885°, E89.10790°, an elevation of 89 m below sea level) in the Turpan Depression, which is the second lowest depression on Earth. The voucher specimen (No. WGXG08226) was deposited in the herpetological collection at Chengdu Institute of Biology, Chinese Academy of Sciences. We took the similar strategy as described previously (Chen et al. Citation2019) to assemble and annotate this nearly complete mitogenome of P. axillaris. It was deposited in GenBank with accession number MK284224.
The nearly complete mtDNA sequence consists of 15,883 bp and contains two ribosomal RNA genes, 13 protein-coding genes (PCGs), and 22 transfer RNA genes (tRNAs). The gene arrangement and composition is identical with the published mitogenome of P. axillaris, which was collected from the Taklimakan Desert (Li et al. Citation2013). The control region consisted of two parts, with one (651 bp) existing between the tRNA-Thr gene and tRNA-Pro gene and another (with 107 bp already sequenced) inserting between the tRNA-Phe and 12S rRNA. Most genes were encoded on heavy strand (H-strand) except for ND6 and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser[UGA], Glu, and Pro). Most of the PCGs were initiated with the typical ATG codon, except for ND1 with ATA, ND2 and ND3 with ATT, ATP8 and ND6 with GTG. Meanwhile, ATP8, ND4L, ND4, and ND5 genes were terminated with TAA as stop codon; ND2 and COI end with AGA; ND1 ends with TAG; ND6 with AGG and the remaining five PCGs end with the incomplete termination codon T.
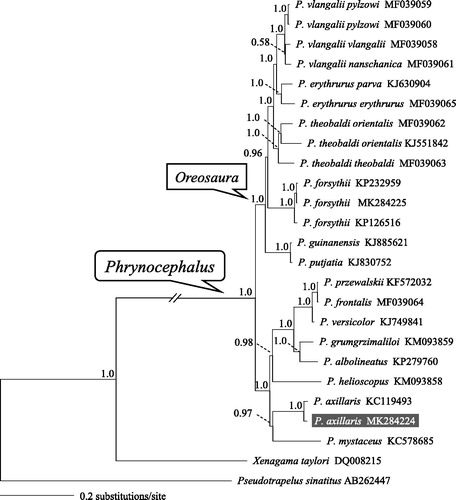
The concatenated PCGs of Phrynocephalus available in GenBank and two Agaminae taxa as outgroups were used to reconstruct the Bayesian phylogenetic tree for assessing mitochondrial sequence authenticity of P. axillaris and its phylogenetic placement. The phylogenetic tree recovered the monophyly of both Phrynocephalus and subgenus Oreosaura (i.e. the viviparous group) (). Two individuals of P. axillaris clustered together and were closely related to P. mystaceus. The newly determined mitogenome sequence will provide fundamental data to investigate altitude-related evolution of mitochondrial genes in Phrynocephalus.
Figure 1. A majority-rule consensus tree inferred from Bayesian inference using MrBayes v.3.2.1 (Ronquist & Huelsenbeck Citation2003) with GTR + г substitution model, based on the concatenated PCGs of 22 individuals of toad-headed lizards and two outgroups. The novel sequencing sample is highlighted. GenBank accession numbers are given with species/subspecies names. DNA sequences were aligned in MEGA v.6.06 (Tamura et al. Citation2013). The PCGs were translated to amino acids sequences and were manually concatenated all sequences into a single nucleotide dataset (total 11,329 bp). Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Chen D, Li J, Guo X. 2019. Next-generation sequencing yields a nearly complete mitochondrial genome of the Forsyth’s toad-headed agama, Phrynocephalus forsythii (Reptilia, Squamata, Agamidae). Mitochondrial DNA B Resour. 4:817–819.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Li J, Guo X, Chen D, Wang Y. 2013. The complete mitochondrial genome of the Yarkand toad-headed agama, Phrynocephalus axillaris (Reptilia, Squamata, Agamidae). Mitochondrial DNA. 24:234–236.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhao E, Zhao K, Zhou K. 1999. Fauna Sinica. Reptilia Vol. 2. Squamata. Lacertilia. Beijing (in Chinese): Science Press.
