Abstract
The complete mitochondrial (mt) genome of Gavia pacifica was sequenced and characterized the circular mt genome, which constituted 37 genes (13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs) and a non-coding region (NCR). Phylogenetic analysis based on the full mt genome sequences confirmed that the Genus Gavia belongs to a monophyletic group containing the G. stellata, G. arctica, and G. pacifica. This is the first completed mt genome from G. pacifica, which provides data for further study of phylogeny in Gaviiformes.
Loons (Gaviiformes) are one of the most successful groups of swimming birds as a specialist foot-propelled diver that includes five extant species: Gavia stellata, G. arctica, G. pacifica, G. immer, and G. adamsii (Boertmann Citation1990; Clifton and Biewener Citation2018). Among loon species, Pacific Loons (Gavia pacifica) reside for the most part along the eastern coast of the Pacific Ocean and Arctic Ocean along Canada’s northern boundary (Birch and Lee Citation1997). The G. pacifica species, previously considered conspecific with the similar black-throated loon (Gavia stellata), was classified as a separate species (AOU Citation1983). Unlike other Loon species, the ecological and biological importance of this species has been poorly understood and particularly, the complete mitochondrial (mt) genome has not been reported. Here, we sequenced the full mt genome of the G. pacifica, which can help its phylogenetic position and evolution of genomes.
The G. pacifica specimen was collected from the southern coast of Korea (34°42′N, 125°25′E). We extracted the genomic DNA from the subsample (muscle) using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol and the extracted DNA sample was deposited at the Wildlife Specimen Bank in Research Center of Ecomimetics, Chonnam National University, Korea. We determined the complete mt genome sequence using the next-generation sequencing reads (400 bp length in each read) generated from MiSeq (Macrogen, Seoul, Korea). Mapped reads were used for de novo assembly and annotation by using commercial software (MITOS, http://mitos.bioinf.uni-leipzig.de/index.py) to identify the full mt genome with about an average 150 × coverage.
The complete mt genome of G. pacifica was 16,178 bp in length deposited in GenBank (G accession No. MK342599) and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes (srRNA and lrRNA), and a putative long non-coding control region (NCR). Twelve protein-coding genes, 14 tRNAs, and 2 rRNAs were predicted to be transcribed from the same strand (heavy strand), whereas 1 protein-coding gene (NADH dehydrogenase subunit 6) and 8 tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu) were encoded on the light strand. The nucleotide composition of the G. pacifica (A = 30.4%, C = 32.2%, G = 14.1%, and T = 23.3%) was similar to that of G. arctica mt genome (A = 30.3%, C = 31.8%, G = 14.2%, and T = 23.7%) and G. stellata (A = 30.4%, C = 31.4%, G = 14.4%, and T = 23.8%). The sequence comparisons between G. pacifica and G. arctica indicated a 95.0% sequence identity, but sequence identity between G. pacifica and G. stellata was 91%, which places the G. stellata sister to these two species ().
Figure 1. Phylogenetic tree of Gavia pacifica and other related species based on complete mitochondrial (mt) genome data. The bootstrap value based on 1000 replicates is shown on each node. Anas platyrhynchos and Pica pica were used as outgroups for tree rooting. The phylogenetic analysis was performed using MEGA7 (Saitou and Nei Citation1987).
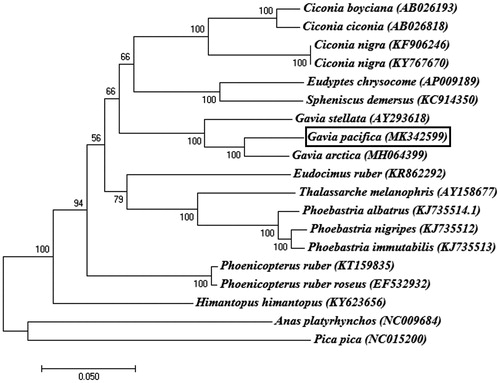
In order to investigate the phylogenetic position of G. pacifica, the full mt genome sequences of 17 Aves species were extracted from GenBank and Anas platyrhynchos and Pica pica served as an outgroup. Consistent with a previous study on phylogenetic analysis with partial mt genome of G. pacifica (Moon et al. Citation2018), phylogenetic analysis based on the full mt genome sequences demonstrated that G. pacifica was clustered in a monophyletic group with other two loon species (G. arctica and G. stellata) and also shared close relationships with Sphenisciformes and Ciconiiformes (). These data provide important molecular data for further evolutionary analysis for the phylogenetic relationships of the family and also a useful genetic marker for identification and ecological studies on Gaviiformes.
Disclosure statement
The authors declare no conflict of interests.
Data availability
GenBank accession number from the complete mitochondrial genome of Gavia pacifica (MK342599) has been registered with the NCBI database.
Additional information
Funding
References
- Birch A, Lee CT. 1997. Field identification of Arctic and Pacific Loons. Birding. 29:106–115.
- Boertmann D. 1990. Phylogeny of the divers, family Gaviidae (Aves.). Steenstrupia. 16:21–36.
- Clifton GT, Biewener AA. 2018. Foot-propelled swimming kinematics and turning strategies in common loons. J Exp Biol. 221.
- American Ornithologists’ Union. 1983. Check-list of North American Birds, 6th ed. American Ornithologists’ Union, Washington, D.C. – 1985. Thirty-fifth supplement to the AOU Check-list of North American Birds. Auk. 102: 680–686.
- Moon JI, Park JG, Hur S, Kim YK, Nam DH, Lee DH. 2018. Mitochondrial genome of the black-throated loon, Gavia arctica (Gaviiformes: Gaviidae): phylogeny and evolutionary history. Mitochondr DNA B. 3:586–587.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
