Abstract
Siniperca kneri is endemic to East Asia with important economic values, and its natural resource is decreasing dramatically. In this study, the complete mitogenome sequence of S. kneri from Hanjiang River was determined using next-generation sequencing. The mitogenome is 16,496 bp in length, including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 non-coding control region. The phylogenetic analyses based on whole mitogenome sequences indicated that S. kneri individuals did not form a monophyly, having a close relationship with Siniperca chuatsi. The present result will provide basic data for the genetic diversity conservation of S. kneri.
Siniperca kneri Garman, commonly named as big-eye mandarin fish, is endemic to East Asia including China and Vietnam (Froese and Pauly Citation2018), having important commercial values because of its high meat quality, appealing taste, relatively fast growth and large size (Fu et al. Citation2017). Due to the construction of hydroelectric projects in the upstream of Hanjiang River, the wild population of S. kneri is decreasing dramatically in the Northwest of China. It is urgent to conserve the natural resources of S. kneri. Analysis of complete mitochondrial genome (mitogenome) sequence may shed light on some genetic background of this species, contributing to the phylogenetic reconstruction of the controversial family Sinipercidae, and could provide basic data for the genetic diversity conservation of this species.
In this study, the complete mitogenome of one S. kneri individual was newly sequenced by Illumina HiSeq4000 (GenBank accession number MK430069). The specimen was collected from Yinghu Lake in Ankang City (32°36'N, 108°51' E), Shaanxi Province, and stored in Qinba Biodiversity Specimen Museum (Voucher specimen: QBSM201711005). Muscles were fixed in 95% ethanol for DNA extraction, and the whole fish was fixed with 10% formalin. Geneious 11.0.2 was used for annotating the newly sequenced mitogenome. Protein-coding genes (PCGs) and rRNAs were annotated by comparing with the published mitogenomes of the Siniperca species (Chen et al. Citation2012).
Totally 47.93 million paired-end reads were obtained for S. kneri, and 4203 reads were used for assembling the complete mitogenome. The mitogenome of S. kneri is 16,496 bp in total length with a coverage of 38.7 X. As other vertebrate mitogenome (Fischer et al. Citation2013; Shi et al. Citation2018; Feng et al. Citation2019), the S. kneri mitogenome is composed of 13 PCGs (cyt b, ATP6, ATP8, COI-III, ND1-6, ND4L), 22 tRNA genes, 2 rRNA genes (12S and 16S rRNA), and 1 control region (CR or D-Loop). Eight tRNAs and ND6 gene are encoded on the L-strand, and the other 29 genes are encoded on the H-strand. Almost all of 13 PCGs start with the regular initiation codon ATG except COI gene with GTG. Six PCGs terminated with the stop codons TAA or TAG. Seven PCGs use incomplete stop codon (TA– or T––). The total length of CR is 836 bp, including three domains: the terminal associated sequence (TAS), the central conserved domain (CD), and the conserved sequence block (CSB), which is congruent with other fishes (Zhao et al. Citation2006; Shi et al. Citation2018).
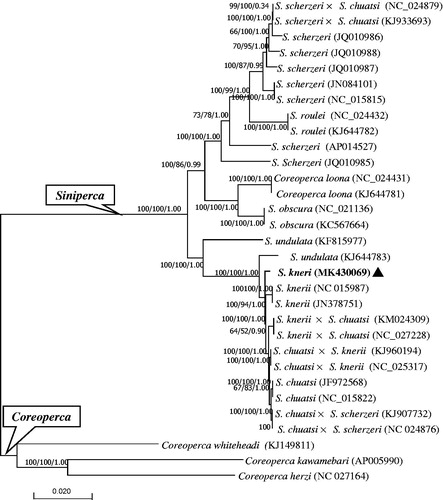
The phylogenetic trees of the family Sinipercidae were reconstructed based on whole mitogenome dataset. Three analysis methods (NJ, ML, and BI) yielded almost identical topologies (). All sinipercid individuals are divided into two clades, corresponding to two genera, Coreoperca and Siniperca, which is congruent with some previous studies (Liu and Chen Citation1994; Zhao et al. Citation2006; Chen et al. Citation2010; Song et al. Citation2017). All hybrids show maternally inherited feature, clustering with their maternal species. Three S. kneri individuals did not form a monophyly, but a paraphyly, and they have a close relationship with Siniperca chuatsi.
Figure 1. Phylogenetic tree of the family Sinipercidae reconstructed using neighbor-joining (NJ), maximum-likelihood (ML), and Bayesian method (BI) based on whole mitogenome sequences. Values at the nodes correspond to the support values for NJ/ML/BI methods. Solid triangle indicates the newly sequenced mitogenome.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen DL, Guo XG, Nie P. 2010. Phylogenetic studies of sinipercid fish (Perciformes: Sinipercidae) based on multiple genes, with first application of an immune-related gene, the virus-induced protein (viperin) gene. Mol Phylogenet Evol. 55:1167–1176.
- Chen DX, Chu WY, Liu XL, Nong XX, Li YL, Du SJ, Zhang JS. 2012. Phylogenetic studies of three sinipercid fishes (Perciformes: Sinipercidae) based on complete mitochondrial DNA sequences. Mitochondrial DNA. 23:70–76.
- Feng X, Chen YX, Sui XY, Chen YF. 2019. The complete mitochondrial genome of Triplophysa sellaefer (Cypriniformes: Balitoridae). Mitochondrial DNA Part B. 4:536–537.
- Fischer C, Koblmüller S, Gülly C, Schlötterer C, Sturmbauer C, Thallinger GG. 2013. Complete mitochondrial DNA sequences of the threadfin cichlid (Petrochromis trewavasae) and the blunthead cichlid (Tropheus moorii) and patterns of mitochondrial genome evolution in cichlid fishes. PLoS One. 8:e67048.
- Froese R, Pauly D. 2018. FishBase. version [2018 June]. www.fishbase.org
- Fu Z, Li J, Shan ST, Huang CC, Ji H, Zeng WX, Chen SW. 2017. Study on the artificial breeding of Siniperca chuatsi using Hypophthalmichthys molitrix feed based on water temperature. Fish Sci Technol Inform. 6:335–339.
- Liu HZ, Chen YY. 1994. Phylogeny of the sinipercine fishes with some taxonomic notes. Zool Res. 15:1–12.
- Shi LX, Zhang C, Wang YP, Tang QY, Danley PD, Liu HZ. 2018. Evolutionary relationships of two balitorids (Cypriniformes, Balitoridae) revealed by comparative mitogenomics. Zool Scr. 47:300–310.
- Song SL, Zhao JL, Li CH. 2017. Species delimitation and phylogenetic reconstruction of the sinipercids (Perciformes: Sinipercidae) based on target enrichment of thousands of nuclear coding sequences. Mol Phylogenet Evol. 111:44–55.
- Zhao JL, Li SF, Cai WQ, Wang WW. 2006. Phylogenetic relationship of sinipercine fishes in East Asia based on cytochrome b sequences analysis. Acta Zool Sin. 4:676–680.
