Abstract
The complete chloroplast genome sequence of Parashorea chinensis, an endangered large tree species in the northern edge of tropical Asia, is determined in this study. The total genome size is 152,002 bp, containing a large single copy (LSC) region (84,094 bp) and a small single copy region (20,003), which were separated by two inverted repeat (IRs) regions (23,954 bp). The overall GC contents of the plastid genome were 37.1%. In total, 116 unique genes were annotated and they consisted of 81 protein-coding genes, 31 tRNA genes, and four rRNA genes. Phylogenetic analysis based on 20 chloroplast genomes indicates that P. chinensis is closely related to P. macrophylla.
Parashorea chinensis is a large tree species (up to 80 m tall) in the family Dipterocarpaceae. It is found in southern China and in northern Vietnam. It has been overexploited and now is threatened by habitat loss (Li et al. Citation2007). The plant is classified as endangered in the IUCN Red List of Threatened Species (Qin et al. Citation2017). Consequently, the genetic and genomic information is urgently needed to promote its systematics research and the development of conservation value of P. chinensis. Here, we made the first report of a complete plastome for P. chinensis (GenBank accession number: MK424049).
The total genomic DNA was extracted from dry leaves collected from Ningming (Guangxi, China, E107.0981°, N22.2456°) and Voucher herbarium specimens were deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden. Genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987). Total DNA was used for the shotgun library construction. After cluster generation, libraries were sequenced on an Illumina Hiseq 4000 platform and 150 bp paired-end reads were generated. The filtered reads were assembled using the program GetOrganelle v1.5 (Jin et al. Citation2018) with the reference chloroplast genome of P. macrophylla (GenBank accession number: MH791330.1), annotated by Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and GeSeq (Tillich et al. Citation2017).
The complete chloroplast genome of P. chinensis is 152,002 base pairs (bp) in length and contains two inverted repeat (IRa and IRb) regions of 23,954 bp, which was separated by a large single-copy (LSC) region of 84,094 bp and a small single-copy (SSC) region of 20,003 bp. The overall GC contents of the plastid genome were 37.1%. The new sequence possesses total 116 genes, including four ribosomal RNA genes, 31 tRNA genes, and 81 protein-coding genes. In these genes, six tRNA genes (i.e. trnE-UUC, trnA-UGC, trnL-UAA, trnS-CGA, trnV-UAC, and trnY-AUA) and seven protein-coding genes (clpP, ndhA, ndhB, rpl2, rps16, rpoC1 and ycf1) contained one intron, and the ycf3 gene have two introns. Most of the genes occurred as a single copy, whereas four rRNA genes (i.e. 4.5S, 5S, 16S, and 23S rRNA), seven tRNA genes (i.e. trnA-UGC, trnE-UUC, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, and trnV-GAC) and six protein-coding genes (i.e. rpl2, rpl23, ycf2, ndhB, rps7, and rps12) occur in double.
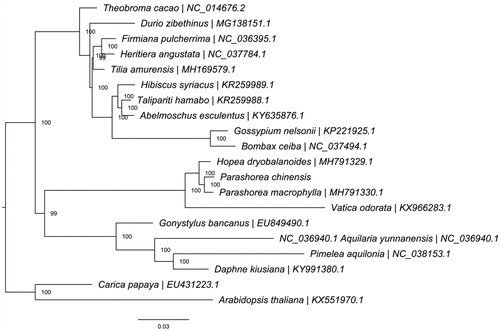
To understand the phylogenetic position of Parashorea within the order Malvales, we downloaded the complete chloroplast genomes of 19 species from the NCBI GenBank database, including 17 species in Malvales and two species in Brassicales. The sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013), and RAxML (Stamatakis Citation2014) was used to construct a maximum likelihood tree with Arabidopsis thaliana and Carica papaya as outgroups. All nodes in the complete plastome trees were strongly supported. The phylogenetic tree showed that P. chinensis was closely related to P. macrophylla (). This published P. chinensis chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Dipterocarpaceae and Malvales.
Figure 1. ML phylogenetic tree of the 18 Malvales based on the available chloroplast genome sequences in GenBank, and the chloroplast sequence of Parashorea chinensis. The tree is rooted with the Brassicales (Arabidopsis thaliana and Carica papaya). Bootstraps (10,000 replicates) are shown at the nodes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Yi T-S, Li D-Z. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li X, Li H-w, Li J, Ashton PS. 2007. Flora of China. Beijing: Science Press. p. 52–54.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, A, et al. 2017. List of threatened species of higher plants in China. Biodiversity. 25:696–744.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq -versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
