Abstract
Huanhu yak is a native yak species of China. In the present study, we reported the complete mitochondrial DNA sequence of the Huanhu yak. The complete mitochondrial genome is a circular molecule with 16,323 bp length which consisted of 13 protein-coding genes, 22 tRNA genes, 2 ribosomal RNA genes, and a non-coding control region (D-loop region). The overall nucleotide composition is 33.71% A, 27.26% T, 25.82% C, and 13.21% G, respectively. The phylogenetic analysis showed that the genetic relationship of Huanhu yak is closer to Pali yak.
Yak (Bos grunniens) belongs to the family of Bovidae, which is endemic to the mountainous areas of Central Asia around the Qinghai-Tibetan Plateau. It provides meat, milk, and transport for the indigenous human population. There are an estimated 14 million yaks in the world, of which 95% are distributed in China (Wiener et al. Citation2003). Huanhu yak is an important yak genetic resource, mainly distributed in the Qinghai lake areas, Qinghai province, China. The typical characteristic of Huanhu yak is the strong capability to adapt to high attitude and better quality meat (Li et al. Citation2016). Mitochondrial DNA has been widely applied as a powerful molecular tool in evolutionary and phylogenetic studies (Stephen Citation2014). In the present study, we assembled and characterized the complete mitochondrial genome of Huanhu yak.
In this study, the specimen of Huanhu yak was obtained from the Haibei Tibetan Autonomous Prefecture Qinghai province, China (N36°57′16.40″, E100°53′58.08″). Muscle tissue samples were frozen in liquid nitrogen and preserved in Key Laboratory of Yak Breeding Engineering of Gansu Province. Total genomic DNA was extracted from the individual muscle tissue samples using the animal tissue DNA isolation kit (ZDGSY, Beijing, China) according to the manufacturer’s instructions. In this study, polymerase chain reaction method was carried out to amplify the whole mitochondrial genome with six pairs of primers. Sequencing results were assembled using DNAStar5.01 software (Madison, WI, USA). The complete mitochondrial genome of Huanhu yak was determined and deposited in GenBank with an accession number: MK124955.
The total length of the mitochondrial DNA is 16,323 bp, with the base composition of 33.71% for A, 27.26% for T, 25.82% for C, and 13.21% for G. Similar to other yak’s mitochondrial structure (Guo et al. Citation2016), it contains the typical structure including 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes (12S and 16S rRNA), and a noncoding control region (D-loop region). Among these genes, NADH dehydrogenase subunit 6 (ND6) and 8 tRNA genes (Gln, Ala, Asn, Cys, Try, Glu, Pro, and Ser) are located on the light strand (L-strand), while all the remaining genes are located on the heavy strand (H-strand).
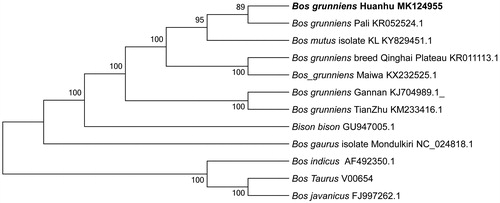
The phylogenetic position of Huanhu yak was reconstructed based on 12 complete mitochondrial genomes of Bovidae using MEGA5.0 software (Tamura et al. Citation2011) with a neighbor-joining (NJ) tree. Bootstrap values were calculated using 1000 replicates to assess node support. The molecular phylogenetic analysis revealed that Huanhu yak has a closer genetic relationship with Pali yak ().
Figure 1. Phylogenetic relationships of 12 species based on protein-coding genes using the neighbor-joining (NJ) methods. The bootstrap values from 1000 replicates are shown next to the branches.
In summary, the complete mitochondrial genome of Huanhu yak will provide essential molecular data for further studies on phylogeny and evolution of yak and this information would be important for evaluation, conservation, and utilization of Huanhu yak.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Guo X, Pei J, Bao P, Chu M, Wu X, Ding X, Yan P. 2016. The complete mitochondrial genome of the Qinghai Plateau yak Bos grunniens (Cetartiodactyla: Bovidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2889–2890.
- Li S, Jing Y, Yan Z. 2016. Experiment of slaughtering Huanhu yaks and meat quality investigation. Food Ind. 7:172–174.
- Stephen L. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wiener G, Han J, Long R. 2003. The Yak. 2nd ed. Regional office for Asia and the Pacific Food and Agriculture Organization of the United Nations. Bangkok, Thailand.
