Abstract
The complete chloroplast genome sequence of Berchemia flavescens was assembled and characterized as a resource for future genetic studies. With a total length of 160,87 bp, the chloroplast genome comprised of a large single-copy (LSC) region of 89,066 bp, a small single-copy (SSC) region of 18,754 bp, and two inverted repeat (IR) regions of 26,526 bp. The overall GC contents of the chloroplast genome were 37.2%. A total of 115 genes were predicted, consisting of 81 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. In these genes, 10 genes contained one intron and two genes contained two introns. Phylogenetic analysis confirmed the position of B. flavescens within the order Rosales.
Berchemia is a genus of plants in the family Rhamnaceae. They are climbing plants or small to medium-sized trees that occur in Africa, Asia, and the Americas. The species B. flavescens occurs naturally in wet shady areas of the central Asian mountains and highlands and are widely found in China, Burhan, and India (Chen and Chou Citation1982). In this study, we characterized the complete chloroplast genome sequence of B. flavescens as a resource for future genetic studies on this and other related species (GenBank accession number: MK460212).
The total genomic DNA was extracted from dry leaves sampled from Tianshui (Gansu, China, E 105.3667°, N 34.6667°, 3654 m a.s.l.) and Voucher herbarium specimens were deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden. Total genomic DNA was extracted with the Qiagen DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA). The genomic paired-end (PE150) sequencing was performed on an Illumina Hiseq 2000 instrument (Illumina, Inc., San Diego, CA, USA). The cp genome was assembled using the program NOVOPlasty (Dierckxsens et al. Citation2016). Annotation was performed using DOGMA (Wyman et al. Citation2004), coupled with manual correction for start and stop codons of protein-coding genes.
The plastome of Berchemia flavescens displayed the typical quadripartite structure and conserved organization as most angiosperms, containing two inverted repeat regions of 26,526 bp, a large single-copy region of 89,066 bp, and a small single-copy region of 18,754 bp. The GC content was 37.2% overall, while higher in IR (42.7%) than in LSC (35.1%) and SSC (31.7%). The plastome comprised 115 unique genes including 81 protein-coding genes (PCGs), 30 tRNAs, and 4 rRNAs. Ten genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16) contained one intron and two genes (ycf3 and clpP) contained two introns. Six protein-coding genes, seven tRNAs, and all four rRNAs were completely duplicated within IRs.
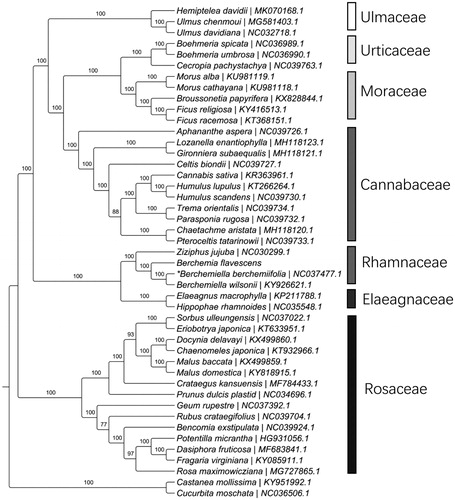
To further investigate its phylogenetic position, a maximum likelihood tree was constructed based on complete chloroplast genome sequences of 42 other Rosales species using RAxML (Stamatakis Citation2014) after the sequences were aligned using MAFFT v7.307 (Katoh and Standley Citation2013). Our results suggested B. flavescens was close to the Berchemiella clade. This published B. flavescens chloroplast genome will provide useful information for phylogenetic and evolutionary studies in Rhamnaceae and Rosales.
Figure 1. Maximum likelihood tree showing the relationship among Berchemia flavescens and representative species within the order Rosales, based on whole chloroplast genome sequences, with Cucurbita moschata (Cucurbitales) and Castanea mollissima (Fagales) as outgroups. Bootstrap support is indicated for each branch based on 1000 replicates. *The species name is Berchemia berchemiifolia in the NCBI, which is the synonym of Berchemiella berchemiifolia (The Plant List Citation2013; Cheon et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chen Y-L, Chou P-K. 1982. Rhamnaceae. Beijing: Science Press.
- Cheon K-S, Kim K-A, Yoo K-O. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA B. 3:133–134.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability . Mol Biol Evol. 30:772–780.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- The Plant List. 2013. Version1.1. Published on the Internet. [accessed 2019 Jan 28]. http://www.theplantlist.org/tpl/record/kew-2674702.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
