Abstract
Liriodendron tulipifera L. is easily recognized by its heart-shaped of leaves and can grow up to 60 meters in North America. In this study, we presented second complete mitochondrial genome of L. tulipifera to understand intra-species variations. Its length is 156,387 bp and contains 66 genes (42 protein-coding genes, three rRNAs, and 21 tRNAs). The overall GC content of the mitochondrial genome is 43.5%, 365 single nucleotide polymorphisms (SNPs; 0.07%), and 2117 insertions and deletions (INDELs; 0.38%) are identified. Most of INDELs are from 1980 bp deletions on this mitochondrial genome. Phylogenetic trees show that intra-species relation in both Liriodendron and Arabidopsis shows are clustered tightly in each clade. In addition, monocots and eudicots are positioned in different manner from classical plant taxonomy, requiring more researches.
Genus Liriodendron L. in Magnoliaceae family contains only two species, which are large deciduous trees (Hunt Citation1998). Liriodendron tulipifera L. is easily recognized by its heart-shaped leaves and can grow up to 60 meters (Strother et al. Citation2015). Liriodendron tulipifera has been studied in various aspects (Cai et al. Citation2006; Liang et al. Citation2008; Jin et al. Citation2011; Liang et al. Citation2011) because it is one of early angiosperms (Chase et al. Citation2016). To date, Amborella truchopoda consisting five chromosomes (Rice et al. Citation2013), Liriodendron tulipifera (Richardson et al. Citation2013), and Nymphaea colorata (Dong et al. Citation2018), which are early angiosperms, are available; but, no mitochondrial genome showing intra-species variations. Total DNA was extracted from fresh leaves of L. tulipifera collected in the expeimental field managed by Kookmin University in Korea using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) to understand intra-species variations. Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea. Mitochondrial genome was completed using Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on L. tulipifera mitochondrial genome (NC_008326) isolated in the United States (Richardson et al. Citation2013).
The mitochondrial genome of Korean L. tulipifera (Genbank accession is MK340747) is 551,806 bp, which is shorter than US L. tulipifera genome by 1,915 bp. It contains 66 genes (42 protein-coding genes, three rRNAs, and 21 tRNAs) and overall GC content is 43.5%, same to those of USA L. tulipifera.
Based on alignment of both Liriodendron mitochondrial genomes, 365 single nucleotide polymorphisms (SNPs; 0.07%) and 2,117 insertions and deletions (INDELs; 0.38%) are identified. 33 INDEL clusters are identified: the largest is 1,980 bp deletion on Korean L. tulipifera genome found in Inverted repeat, accounting for length difference between two genomes. 222 SNPs (60.8%) are in the five regions from its chloroplast genomes, indicating that they were integrated into mitochondrial genomes but not used functionally. All sequence variations are identified outside of coding regions, tRNAs, and rRNAs, which is similar to the case of Arabidopsis thaliana (180404IB4; Korean natural isolate; Park et al., in preparation).
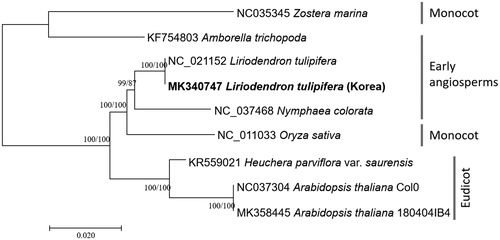
Nine complete mitochondrial genomes including two Liriodendron (Richardson et al. Citation2013), Amborella (Rice et al. Citation2013) and Nymphaea (Dong et al. Citation2018), as well as two Arabidopsis mitochondrial genomes (Sloan et al. Citation2018; Park et al., in preparation) to compare intra-species variations were used for drawing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) trees based on alignments of 13 common protein-coding genes using MAFFT 7.388 (Katoh and Standley Citation2013) and MEGA X (Kumar et al. Citation2018). Phylogenetic trees show that intra-species relation in both Liriodendron and Arabidopsis shows are clustered tightly in each clade because of low sequence variations (). Trees also show that eudicot clade is not nested in monocot and early angiosperm clades and two monocot species are not clustered in one clade, disagreeing with classical plant taxonomy (), requiring additional in-depth analyses of these mitochondrial genome sequences.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of nine complete mitochondrial genomes: two Liriodendron tulipifera (MK340747 in this study and NC_021152), Amborella trichopoda (KF754803), Nymphaea colorata (NC_037468), Heuchera parviflora var. saurensis (KR559021), Zostera marina (NC_035345), Oryza sativa (NC_011033), and Arabidopsis thaliana Col-0 and 180404IB4 (NC_037304 and MK358445). Phylogenetic tree was drawn based on neighbor joining tree. Grey bars indicate specific clades with labels. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, dePamphilis CW, Boore JL, Jansen RK. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77.
- Chase MW, Christenhusz M, Fay M, Byng J, Judd WS, Soltis D, Mabberley D, Sennikov A, Soltis PS, Stevens PF. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Dong S, Zhao C, Chen F, Liu Y, Zhang S, Wu H, Zhang L, Liu Y. 2018. The complete mitochondrial genome of the early flowering plant Nymphaea colorata is highly repetitive with low recombination. BMC Genom. 19:614.
- Hunt D. 1998. Magnolias and their allies. Published for the International Dendrology Society and the Magnolia Society.
- Jin H, Do J, Moon D, Noh EW, Kim W, Kwon M. 2011. EST analysis of functional genes associated with cell wall biosynthesis and modification in the secondary xylem of the yellow poplar (Liriodendron tulipifera) stem during early stage of tension wood formation. Planta. 234:959.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Liang H, Ayyampalayam S, Wickett N, Barakat A, Xu Y, Landherr L, Ralph PE, Jiao Y, Xu T, Schlarbaum SE. 2011. Generation of a large-scale genomic resource for functional and comparative genomics in Liriodendron tulipifera L. Tree Genet Genom. 7:941–954.
- Liang H, Carlson JE, Leebens-Mack JH, Wall PK, Mueller LA, Buzgo M, Landherr LL, Hu Y, DiLoreto DS, Ilut DC. 2008. An EST database for Liriodendron tulipifera L. floral buds: the first EST resource for functional and comparative genomics in Liriodendron. Tree Genetics & Genomes. 4:419–433.
- Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, et al. 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 342:1468–1473.
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. 2013. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 11:29.
- Sloan DB, Wu Z, Sharbrough J. 2018. Correction of persistent errors in Arabidopsis reference mitochondrial genomes. Plant Cell. 30:525–527.
- Strother CW, Madden M, Jordan TR, Presotto A. 2015. Applications paper: Lidar detection of the ten tallest trees in the Tennessee portion of the Great Smoky Mountains National Park. Photogramm Eng Remote Sens. 81:407–413.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
