Abstract
The complete chloroplast genome of Archakebia apetala, a monotypic genus endemic to China, was determined. The size of complete chloroplast genome is 157,929 bp, containing a large single copy (LSC) region of 86,630 bp and a small single copy region (SSC) of 19,001 bp, separated by a pair of inverted repeats (IRs) of 26,149 bp. The chloroplast genome encodes 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Among them, 15 genes have one intron each and 3 genes contain two introns. The overall GC content is 38.7%, while the corresponding values of LSC, SSC, and IR regions are 37.1, 33.6, and 43.1%, respectively. Phylogenetic relationship analysis showed that Archakebia apetala was closely related to Holboellia latifolia. The complete chloroplast genome could provide new insight into the evolution of Lardizabalaceae.
Archakebia C. Y. Wu (Lardizabalaceae) is a monotypic genus endemic to China, with the species, Archakebia apetala (Q. Xia, J. Z. Suen & Z. X. Peng) C. Y. Wu, distributed in the narrow regions of Gansu, Sichuan, and Shaanxi provinces (Wu et al. Citation2001). The genus Archakebia is similar to the genus Akebia in many characteristics such as stem, leaf, fruit, and flower, except sepals 6, lanceolate or linear, which are common in the genus Stauntonia (Qin Citation1995). It is generally considered that Archakebia is sister to Akebia, but molecular evidence is still lacking. As complete chloroplast genomes can offer valuable information for the reconstruction of complicated evolutionary relationships, they have been widely used for phylogenetic reconstruction in recent years (Raman and Park Citation2015). In this study, we reported and characterized the complete chloroplast genome sequence of Archakebia apetala.
Fresh leaves were collected from a single individual planted in the Germplasm nursery of Akebia in Jiujiang University and the resource was gathered from Wen county, Gansu province, China (32.51.806N, 104.24.249E). The total genomic DNA was extracted using the modified CTAB method (Doyle Citation1987). The genomic library was sequenced on an Illumine Hiseq X Ten platform (Shanghai, China) with paired-end reads of 150 bp. The chloroplast genome was assembled via BBMap (Bushnell Citation2014) implemented in Geneious (Kearse et al. Citation2012), using the Akebia trifoliata chloroplast as a reference (GenBank accession NC_029427). The finished chloroplast genome was annotated using the Geneious ‘Annotate from Database’ tool (Kearse et al. Citation2012). Finally, the annotated complete chloroplast genome of Archakebia apetala was submitted to GenBank (accession MK468518).
The size of complete chloroplast genome of Archakebia apetala is 157,929 bp with high coverage (mean 1431×). It has a typical quadripartite structure, including a large single copy (LSC) region of 86,630 bp, a small single copy region (SSC) of 19,001 bp, and a pair of inverted repeats (IRs) of 26,149 bp. There are 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, consistent with the chloroplast genome of Akebia. Among these genes, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) have one intron each, and three genes (clpP, rps12 and ycf3) contain two introns. The overall GC content is 38.7%, while the GC content of LSC, SSC, and IR regions are 37.1, 33.6, and 43.1%, respectively.
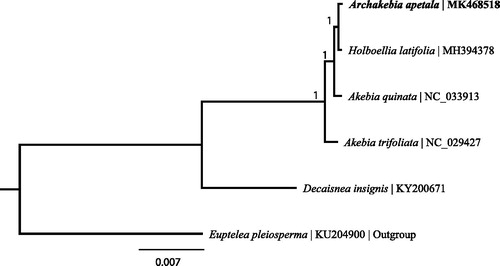
The phylogenetic tree including Archakebia apetala, four Lardizabalaceae species, and the outgroup Euptelea pleiosperma was constructed by complete chloroplast genomes. The genome-wide alignment was carried out by MAFFT (Katoh and Standley Citation2013). Also, the phylogenetic tree was generated by MrBayes (Huelsenbeck and Ronquist Citation2001). In contrast to previous findings (Wang et al. Citation2002; Wang et al. Citation2009), Archakebia apetala is most closely related to Holboellia latifolia (). The chloroplast genome determined in this study could provide essential data to determine the phylogenetic position of Archakebia apetala and provide new insight into the evolution of Lardizabalaceae.
Additional information
Funding
References
- Bushnell B. 2014. BBMap: A fast, accurate, splice-aware aligner. Berkeley, CA (US): Ernest Orlando Lawrence Berkeley National Laboratory.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:647–1649.
- Qin HN. 1995. Archakebia – a new genus of Lardizabalaceae from China. Acta Phytotaxon Sin. 33:240–243.
- Raman G, Park S. 2015. Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. Plos One. 10:e0141329.
- Wang F, Li DZ, Yang JB. 2002. Molecular phylogeny of the Lardizabalaceae based on TrnL-F sequences and combined chloroplast data. Acta Phytotaxon Sin. 44:971–977.
- Wang W, Lu AM, Endress ME, Chen Z, Ren Y. 2009. Phylogeny and classification of Ranuncululales: evidence from four molecular loci and morphological data. Perspect Plant Ecol. 11:81–110.
- Wu ZY, Raven PH, Hong DY, editors. 2001. Flora of China. Vol. 6. In: Caryophyllaceae through Lardizabalaceae. Beijing: Science Press.