Abstract
Sinojackia sarcocarpa is an endangered plant endemic to China. Here, the complete chloroplast genome of S. sarcocarpa was sequenced and characterized. The total genome size is 158,737 bp, with a typical quadripartite structure including a pair of inverted repeat regions (IRs, 26,090 bp) separated by a large single copy (LSC, 88,002 bp) region and a small single copy (SSC, 18,555 bp) region. The genome contains 137 genes, including 90 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. Phylogenetic analysis revealed that S. sarcocarpa is closely related to Sinojackia xylocarpa and Sinojackia rehderiana, but forms an independent evolutionary branch.
Sinojackia sarcocarpa is a Styracaceae plant endemic to Sichuan province, China (Luo Citation1992). Sinojackia sarcocarpa has good ornamental value and can be used as a garden tree species. In addition, it contains effective anticancer compounds and can be used as an important plant source for the extraction of drug active substances (Wang et al. Citation2011). However, the number of existing wild individuals is very small, S. sarcocarpa is, therefore, under state protection (category II) in China. A good knowledge of its phylogeny and taxonomy is necessary for the protection and utilization of S. sarcocarpa. Molecular systematics has become an important method of taxonomic research. In the past, system evolution was usually inferred by selecting individual or several genes, however, single-gene data lacks sufficient genetic information when compared to the multi-gene data (Thorne and Kishino Citation2002). The chloroplast genome structure is conservative, rarely recombinant, and mutated, and is uniparental inheritance in most cases, so it is of great practical value to study the evolution of plant system (He et al. Citation2019). However, at present, no complete chloroplast genome sequence of S. sarcocarpa is available in the NCBI GenBank. This work is the first to report on the chloroplast genome sequence of S. sarcocarpa, which can better understand the genetic background of S. sarcocarpa and contribute to the conservation of the species.
The samples of S. sarcocarpa were collected from botanical garden in Leshan, Sichuan Province, China and the specimens were stored in the herbarium of Leshan normal university. Total genomic DNA was extracted from 50 mg fresh leaves using SDS method. The complete chloroplast genome of S. sarcocarpa was sequenced by Illumina HiSeq Xten platform, the Illumina raw sequence reads were edited by the NGS QC tool Kit v2.3.3, the percentage of cut-off values for read length and phred quality score were 80 and 30, respectively (Patel and Jain Citation2012). High-quality readings were assembled by SPAdes3.9.0 software, and the K-mer set values were selected as 93, 105, 117, 121 (Bankevich et al. Citation2012). Annotations were performed with the plann software (Huang and Cronk Citation2015). The complete chloroplast genome of S. sarcocarpa (GenBank accession number MK351986) is 158,737 bp in length, which has a GC content of 37.24%, including a large single-copy area (LSC, 88,002 bp) and a small single-copy area (SSC, 18,555 bp), as well as two reverse repetition zones (IRs) of 26,090 bp. The complete chloroplast genome encodes 137 genes, including 90 protein-coding genes, 8 ribosomal RNA genes, and 38 transfer RNA genes, 15 genes contain an intron, 2 genes contain two introns, and 1 rps12 gene has trans splicing.
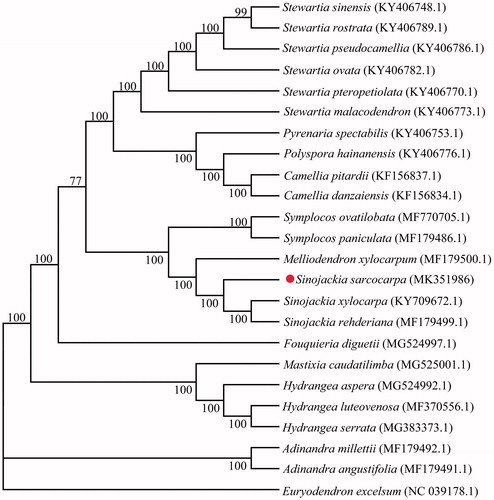
To ascertain the phylogenetic evolution of S. sarcocarpa, the fasta format file containing all the chloroplast genome sequences of 24 plants was treated by MAFFT 7.037 software and then aligned in automatic mode (Katoh and Standley Citation2013). The neighbour-joining (NJ) phylogenetic tree was constructed by MEGA 6.0 software (Tamura et al. 2013), the parameter settings were as follows: the stability of branches was evaluated with 1,000 bootstrap replications, nucleotide substitution model was computed using the p-distance method, the substitution included a variety of transitions and transversions, and the gaps/missing data treatments were set to complete deletion. The results of phylogenetic analysis show that S. sarcocarpa forms a separate branch when compared with other plants in this study, which is more closely related to two other Sinojackia species (). The complete plastome sequence of S. sarcocarpa provides an important data set for the conservation genetics of this species.
Disclosure statement
The authors confirm this article content has no conflict of interest and all the authors are responsible for the content of this article.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- He P, Ma Q, Dong M, Yang Z, Liu L. 2019. The complete chloroplast genome of Leontice incerta and phylogeny of Berberidaceae. Mitochondrial DNA B. 4:101–102.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Luo LQ. 1992. A new species of Sinojackia from Sichuan. Acta Sci Nat Univ Sunyatseni. 31:78–79.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Thorne JL, Kishino H. 2002. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 51:689–702.
- Wang O, Liu S, Zou J, Lu L, Chen L, Qiu S, Li H, Lu X. 2011. Anticancer activity of 2α, 3α, 19β, 23β-tetrahydroxyurs-12-en-28-oic acid (THA), a novel triterpenoid isolated from Sinojackia sarcocarpa. PLoS One. 6:e21130.