Abstract
New candidate of variety of Potentilla, named as Potentilla freyniana var. chejuensis, was found in about 1,000–1,500 m elevations in Mt. Halla in Korea. To uncover its phylogenomic relationships with available chloroplast genomes, we presented complete chloroplast genome of P. freyniana var. chejuensis which is 156,739 bp long and has four subregions: 85,909 bp of large single copy (LSC) and 18,590 bp of small single copy (SSC) regions are separated by 25,940 bp of inverted repeat (IR) regions including 128 genes (84 protein-coding genes, eight rRNAs, and 36 tRNAs). The overall GC content of the chloroplast genome is 36.9% and those in the LSC, SSC, and IR regions are 34.7, 30.7, and 42.8%, respectively. Phylogenomic trees based on whole chloroplast genomes show that P. freyniana var. chejuensis is clustered with three Potentilla species isolated in Korea which present enough differences of their morphologies. It indicates that P. freyniana var. chejuensis can be a new variety species.
Most of around 500 species in genus Potentilla L. (Soják Citation2008) distribute in a Northern Hemisphere. Especially, Asia is a centre of species diversity of Potentilla (Dobeš and Paule Citation2010). Around 30 Potentilla species have been identified in Korea (Heo et al. under revision). We found a new candidate variety species of Potentilla distributed in about 1,000–1,500 m elevations in Mt. Halla where has unique biogeographic feature and endemic species of about 53 ∼ 89 species (Nakai Citation1914; Lee Citation1985; Kim Citation2009; Kong et al. Citation2014). This species is similar with P. freyniana Bornm. of section Potentilla (Syme) Yü et Li, which does not develop stolon or short slender stolon and plant is ca. 8–25 cm tall (Li et al. Citation2003; Naruhashi Citation2003); while it shows very developed in stolon and the plant size is small (Heo et al. in preparation). To clarity phylogenomic relationship of candidates for new Potentilla species named as P. freyniana var. chejuensis; its chloroplast genome was sequenced and analysed.
Total DNA of P. freyniana var. chejuensis collected in Jeju island, Republic of Korea (Voucher in InfoBoss Cyber Herbarium (IN); K-I. Heo, IB-00592; InfoBoss Co., Ltd., Seoul, Korea) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Seoul, Korea, and de novo assembly and sequence confirmations were done by Velvet version 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on Potentilla centigrana chloroplast complete genome (MK209637; Park, Heo et al. Citation2019).
The chloroplast genome of P. freyniana var. chejuensis (Genbank accession is MK472813) is 156,739 bp (GC ratio is 36.9%) and has four subregions: 85,909 bp of large single copy (LSC; 34.7%) and 18,590 bp of small single copy (SSC; 30.7%) regions are separated by 25,940 bp of inverted repeat (IR; 42.8%). It contained 128 genes (84 protein-coding genes, eight rRNAs, and 36 tRNAs); 16 genes (5 protein-coding genes, 4 rRNAs, and 7 tRNAs) are duplicated in IR regions.
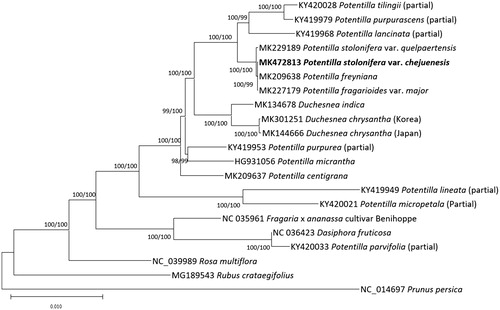
Thirteen partial or complete Potentilla (Ferrarini et al. Citation2013; Zhang et al. Citation2017; Park, Kim et al. Citation2019; Park, Heo et al. Citation2019), three Duchesnea (Heo et al. Citation2019; Park, Kim et al. Citation2019; https://www.tandfonline.com/doi/full/10.1080/23802359.2019.1573114), and five Rosaceae chloroplast genomes (Cheng et al. Citation2017; Yang et al. Citation2017; Zhao et al. Citation2018) were used for constructing neighbour joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees using MEGA X (Kumar et al. Citation2018) based on sequence alignment of whole chloroplast genomes by MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees clearly show that P. freyniana var. chejuensis is clustered with three Potentilla species isolated in Korea which present enough differences of their morphologies (), and it has enough genetic distances among four Potentilla species. It indicates that P. freyniana var. chejuensis can be a new variety of P. freyniana in the aspect of phylogenomic relationship based on whole chloroplast genome sequences.
Figure 1. Neighbour joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 18 Rosaceae partial or complete chloroplast genomes: Potentilla sp. (MK472813, in this study), Potentilla stolonifera var. quelpaertensis (MK229189), Potentilla stolonifera (MK227179), Potentilla freyniana (MK209638), Potentilla centigrana (MK209637), Duchesnea chrysantha (MK182726 and MK301251), Duchesnea indica (MK134678), Potentilla tilingii (KY420028; partial genome), Potentilla lancinata (KY419968; partial genome), Potentilla purpurea (KY419953; partial genome), Potentila purpurascens (KY419979; partial genome), Potentilla micropetala (KY420021; partial genome), Potentilla micrantha (HG931056), Potentilla lineata (KY419949; partial genome), Dasiphora fruticosa (NC 036423), Fragaria x ananassa cultivar Benihoppe (NC_035961), Rubus crataegifolius (MG189543), Rosa multiflora (NC_039989), and Prunus persica (NC_014697). Phylogenetic tree was drawn based on neighbour joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbour joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cheng H, Li J, Zhang H, Cai B, Gao Z, Qiao Y, Mi L. 2017. The complete chloroplast genome sequence of strawberry (Fragaria x ananassa Duch.) and comparison with related species of Rosaceae. PeerJ. 5:e3919.
- Dobeš C, Paule J. 2010. A comprehensive chloroplast DNA-based phylogeny of the genus Potentilla (Rosaceae): implications for its geographic origin, phylogeography and generic circumscription. Mol Phylogenet Evol. 56:156–175.
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, et al. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genom. 14:670.
- Heo KI, Kim Y, Maki M, Park J. 2019. The complete chloroplast genome of mock strawberry, Duchesnea indica (Andrews) Th. Wolf (Rosoideae). Mitochondrial DNA B. 4:560–562.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim CS. 2009. Vascular plant diversity of Jeju island, Korea. Korean J Plant Resour. 22:558–570.
- Kong WS, Kim K, Lee S, Park H, Cho SH. 2014. Distribution of high mountain plants and species vulnerability against climate change. J Environ Impact Assess. 23:119–136.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lee TB. 1985. Endemic and rare plants of Mt. Halla.
- Li C, Ikeda H, Ohba H, Zhengyi W, Raven P, Hong D. 2003. Potentilla L. Flora Chin. 9:291–328.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Nakai T. 1914. Flora of Quelpaert and Wando Island. Seoul, South Korea: Govern Chosen.
- Naruhashi N. 2003. Rosoideae in Flora of Japan. Vol. IIb. Tokyo, Japan: Kodansha.
- Park J, Heo KI, Kim Y, Kwon W. 2019. The complete chloroplast genome of Potentilla centigrana Maxim. (Rosaceae). Mitochondrial DNA B. 4:688–689.
- Park J, Kim Y, Lee K. 2019. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. & Moritzi) Miq. (Rosoideae). Mitochondrial DNA B. 4:864–865.
- Soják J. 2008. Notes on Potentilla XXI. A new division of the tribe Potentilleae (Rosaceae) and notes on generic delimitations. Bot Jahrb. 127:349–358.
- Yang JY, Pak JH, Kim SC. 2017. The complete chloroplast genome sequence of Korean raspberry Rubus crataegifolius (Rosaceae). Mitochondrial DNA B. 2:793–794.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genom Res. 18:821–829.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytologist. 214:1355–1367.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
- Zhao Y, Lu D, Han R, Wang L, Qin P. 2018. The complete chloroplast genome sequence of the shrubby cinquefoil Dasiphora fruticosa (Rosales: Rosaceae). Conserv Genet Resour. 10:675–678.
