Abstract
Alternanthera philoxeroides is an invasive amphibious herbaceous clonal plant. The chloroplast (cp) genome of A. philoxeroides is 151,972 bp long. It encodes a set of 111 unique genes (including 78 protein coding, 29 tRNA, and four rRNA genes). Besides, 11 genes (atpF, ndhA, ndhB, rpoC1, rps12, rps16, trnA-TGC, trnI-GAT, trnK-TTT, trnL-TAA, and trnV-TAC) possess a single intron, and another two genes (clpP and ycf3) have double introns. A neighbour-joining (NJ) phylogenetic analysis revealed that the cp genome of A. philoxeroides is closely related to that of Amaranthus tricolor, Amaranthus hybridus, Amaranthus caudatus, and Amaranthus hypochondriacus.
Alternanthera philoxeroides (Mart.) Griseb (alligator weed) is an invasive amphibious herbaceous clonal plant (Amaranthaceae), which originated in South America, but has now invaded into many aquatic or semi-aquatic situations in 32 different countries across the world (Clements et al. Citation2011; Tanveer et al. Citation2018). In invaded water bodies, this plant grows and reproduces rapidly within a small span of time, forms dense tangled monospecific stands with a large number of underground rhizomatous and roots, excludes native flora and fauna, threats to the biodiversity, changes the structure and function of the surrounding ecosystem, and causes great environmental and economic problems (Bassett et al. Citation2012; Chen et al. Citation2013; Yu and Fan Citation2018). The chloroplast (cp) genome information is a valuable resource for plant phylogenetics and population genetics research and may provide valuable guidelines for the management of A. philoxeroides. In this study, we have mapped and determined the complete chloroplast genome sequence of A. philoxeroides based on the high-throughput Illumina sequencing technology.
Fresh leaves were collected from a single individual of A. philoxeroides in Shiyan City, Hubei Province of China (110°39′59′′E, 32°50′55″) with voucher specimen also deposited at the Anqing Normal University Herbarium (AQ2018101703), and were used for the subsequent genomic DNA extraction with the DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). High-throughput DNA sequencing was conducted on the Illumina HiSeq X Ten Sequencing System (Illumina, San Diego, CA). Approximately, 6.5 Gb of sequence data were generated and used for the assembly of cp genome with MITObim version 1.9 (https://github.com/chrishah/MITObim) (Hahn et al. Citation2013). The cp genome of Amaranthus hypochondriacus (KX279888) (Chaney et al. Citation2016) was included as the initial reference. Genome annotation was conducted in Geneious R10 (Biomatters Ltd., Auckland, New Zealand) by aligning with those of phylogenetically related species.
The complete chloroplast genome sequence of A. philoxeroides (GenBank accession MK 450441) was 151,972 bp in length, with a large single-copy (LSC) region of 84,387 bp, a small single-copy (SSC) region of 17,343 bp, and two inverted repeat (IR) regions of 25,121 bp each. The overall AT content was 63.60%. A total of 111 genes were predicted, consisting of 78 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Out of the 111 genes, the occurrence of double introns was detected in two genes (clpP and ycf3), and the occurrence of one intron in 11 genes (atpF, ndhA, ndhB, rpoC1, rps12, rps16, trnA-TGC, trnI-GAT, trnK-TTT, trnL-TAA, and trnV-TAC).
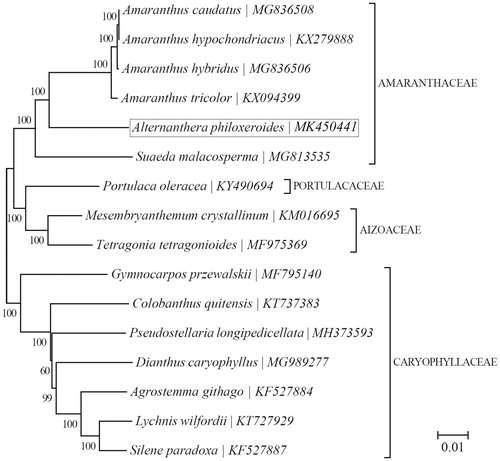
To further investigate its phylogenetic position, a neighbour-joining (NJ) phylogeny was constructed based on complete chloroplast genome sequences of 15 other species within the core Caryophyllales using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates. Phylogenetic analysis showed that A. philoxeroides clustered together with Amaranthus tricolor, Amaranthus hybridus, Amaranthus caudatus, and A. hypochondriacus with 100% bootstrap support values (). The chloroplast resource may provide valuable guidelines for the management of A. philoxeroides.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Bassett I, Paynter Q, Hankin R, Beggs JR. 2012. Characterising alligator weed (Alternanthera philoxeroides; Amaranthaceae) invasion at a northern New Zealand lake. New Zeal J Ecol. 36:216–222.
- Chaney L, Mangelson R, Ramaraj T, Jellen EN, Maughan PJ. 2016. The complete chloroplast genome sequences for four Amaranthus species (Amaranthaceae). Appl Plant Sci. 4:1600063.
- Chen Y, Zhou Y, Yin TF, Liu CX, Luo FL. 2013. The invasive wetland plant Alternanthera philoxeroides shows a higher tolerance to waterlogging than its native congener Alternanthera sessilis. PLoS One. 11:e81456.
- Clements D, Dugdale TM, Hunt TD. 2011. Growth of aquatic alligator weed (Alternanthera philoxeroides) over 5 years in south-east Australia. Aquat Invasions. 6:77–82.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Tanveer A, Ali HH, Manalil S, Raza A, Chauhan BS. 2018. Eco-biology and management of Alligator Weed [Alternanthera philoxeroides)(Mart.) Griseb.]: a review. Wetlands. 38:1067–1079.
- Yu H, Fan S. 2018. Differences in physiological traits and resistances of Alternanthera philoxeroides after herbivory by generalists and specialists. Aquat Ecol. 52:323–332.