Abstract
Haemadipsa japonica is the most common land leech species found in Japan. It has been considered to possess genetic variation that limits its habitat range. In the present study, to characterize variation in the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene of H. japonica specimens from various locations in Japan, we examined nucleotide sequences of the mitochondrial cox1 gene. We performed PCR of mitochondrial cox1 using 10 H. japonica specimens and compared the result with those of land leeches from around the world using a maximum likelihood (ML) tree. ML tree of H. japonica in Japan showed significant differences between cox1 sequences of specimens from Yakushima and other regions of Japan. ML tree of land leeches from around the world revealed that H. japonica had the closest relationship with H. picta from Borneo.
Haemadipsa japonica (H. japonica) is a common hematophagous land leech found in Japan. H. japonica has evolved to be locally distributed. H. japonica specimens found in various regions of Japan lack morphological differences and cannot thus be classified based on morphological features. However, they are thought to possess genetic variation that limits their habitat range. To assess phylogeographic patterns in H. japonica located at sites across Japan, we examined nucleotide sequences of the cox1 gene of mitochondrial DNA (mtDNA).
To determine the species identity and evaluate the genetic relationships among H. japonica specimens throughout Japan, cox1 gene sequences were amplified from 10 H. japonica specimens. Specimens were collected from along bushes and the forest floor in Yakushima (Kagoshima) (30.452222N, 130.485277E), Mie (35.196944N, 136.543611E), Shiga (35.344444N, 135.922777E), Kanagawa (35.512777N, 139.247499E), Shizuoka (35.586946N, 138.314166E), Gunma (36.704722N, 138.986388E), Kimitsu (Chiba) (35.204445N, 140.1405580E), Gifu (35.527499N, 136.639722E), Kamogawa (Chiba) (35.134444N, 140.150277E), and Akita (39.796668N, 140.235001E).
mtDNA was extracted using phenol/chloroform method. Two primers (5’-ATT AAG AGG AAT ACC GCC ATT AAC TGG-3’ for forward primer and 5’-AAT AGC AGG TGC AAT TGT TCA TAA AGT TTC-3’ for reverse primer) for amplification and five primer sets for sequencing of mtDNA were designed. After long PCR using Pfu-x DNA polymerase (Greiner Bio-One, Frickenhausen, Germany), PCR products (2064 bp) were sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, CA, USA) with the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Complete cox1 sequence of H. japonica at Kanagawa was 1569 bp long. This sequence has been submitted to GenBank (accession no. LC424190) as a representative.
Multiple comparisons were performed using MEGA software version 7 to evaluate levels of similarity and differences between nucleotide sequences (Hall Citation2013). A phylogenetic tree obtained by combining our data with previous data (604 bp) (Lai et al. Citation2011; Won et al. Citation2014) is shown in . Maximum likelihood (ML) estimation determines values for parameters of a statistical model such that the parameter values maximize the likelihood that the process described by the model was produced by the observed data. Using the 604 bp cox1 sequences, ML tree showed that H. japonica in Japan belonged to a distinct group from other Haemadipsa. ML tree of land leeches from among the world revealed that H. japonica had the closest relationship with H. picta from Borneo.
Figure 1. Maximum likelihood (ML) tree based on mitochondrial cox1 sequences. (A) ML tree based on 604 bp cox1 sequences of land leech specimens from around the world and 10 H. japonica specimens from Japan. (B) ML tree based on 1569 bp cox1 sequences of 10 H. japonica specimens from Japan. The bootstrap value based on 1000 replicates is shown on each node.
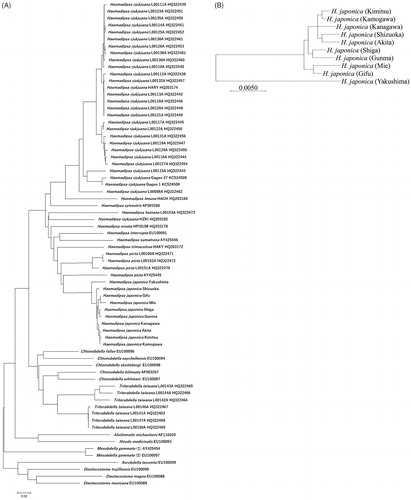
Moreover, ML tree showed a significant difference between complete cox1 sequences (1569 bp) of H. japonica at Yakushima and other regions of Japan (). Furthermore, our findings suggest that H. japonica inhabiting Honshu Island can be separated into four major haplotypes.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 30:1229–1235.
- Lai YT, Nakano T, Chen JH. 2011. Three species of land leeches from Taiwan, Haemadipsa rjukjuana comb. n., a new record for Haemadipsa picta Moore, and an updated description of Tritetrabdella taiwana (Oka). ZooKeys. 139:1–22.
- Won S, Park BK, Kim BJ, Kim HW, Kang JG, Park TS, Seo HY, Eun Y, Kim KG, Chae JS. 2014. Molecular Identification of Haemadipsa rjukjuana (Hirudiniformes: Haemadipsidae) in Gageo Island, Korea. Korean J Parasitol. 52:169–175.
