Abstract
Chaetognaths possess simplified body plan that puzzled their morphological phylogenetic classification. Three complete mitochondrial genomes were determined for Zonosagitta bedoti, Z. pulchra, and Aidanosagitta regularis, and one partial mitochondrial genome was obtained for Ferosagitta robusta. The mitochondrial genomes of the species were small owing to the loss of 23 genes, ranging from 11,604 bp to 12,123 bp. Large-scale rearrangement was also observed in the obtained mitochondrial genomes resembled those of most other Sagittidae species. Our phylogenetic results questioned monophylic status of Zonosagitta because of the inclusion of Ferosagitta and Aidanosagitta in the Zonosagitta clade. Monophyly of Parasagitta and Feerosagitta were supported.
Introduction
Chaetognaths (arrow worms) are common holoplankton species living in epipelagic, mesopelagic, and abyssal layers of the world’s oceans, showing low speciation and displaying a simple body plan (Ball and Miller Citation2006; Gasmi et al. Citation2014). As the dominant holopankton species in the world’s ocean, they play important roles in the food web and carbon flux in the marine ecosystem (Giesecke et al. Citation2010; Sun et al. Citation2010). Simplistic morphological characters of Chaetognaths complicate species identifications (Thuesen et al. Citation1993) and their phylogenetic classification (Bieri Citation1991; Halanych Citation1996; Ball and Miller Citation2006). Nowadays, more than 120 arrow worms have been identified in the world, and their internal systematic relationships remain controversial (Faure and Casanova Citation2006; Yvan et al. Citation2014).
The phylogenetic relationships of sagittidae seem to be problematic (Bieri Citation1991). Many revisions have been proposed in the internal systematic relationships within Sagittidae (Ball and Miller Citation2006; Miyamoto et al. Citation2010; Gasmi et al. Citation2014). It is now considered composed of 12 genera, including Aidanosagitta, Caecosagitta. Decipisagitta, Ferosagitta, Flaccisagitta, Mesosagitta, Parasagitta, Pseudosagitta, Sagitta, Serratosagitta, Solidosagitta, and Zonosagitta (http://www.marinespecies.org/). However, few molecular works are available to confirm their validity and to interpret their phylogenetic interactions (Gasmi et al. Citation2014). Furthermore, Chaetognath mt DNAs was one of the smallest mtDNAs of metazoan (Gasmi et al. Citation2014; Barthélémy and Seligmann Citation2016; Shen et al. Citation2016; Wei et al. Citation2016; Marlétaz et al. Citation2017; Wei et al. Citation2019). Two ATP6/8 genes, as well as majority tRNAs, were found lost in the available arrow worms’ mitochondrial genomes. We would like to find whether the size reduction of mitochondrial genomes is a common phenomenon in the taxon. We sequenced mitochondrial genomes of four arrow worms to shed light on their phylogenetic frameworks and evolutionary details.
Materials and methods
The four Sagittidae specimens used in this study were collected using vertical net-towing (500 μm) during a cruise during Aug 2016 voyage by scientific RV Kexue 3. Specimen of Zonosagitta bedoti, Zonosagitta pulchra, Aidanosagitta regularis, and Ferosagitta robusta were sampled in the East China Sea (Station 1, 122.420°E, 27.051°N; Station 2, 121.074°E, 26.014°N; Station 3, 122.030°E, 26.470°N; Station 4, 120.775°E, 26.343°N). All samples were identified to species level immediately on board and preserved in absolute ethyl alcohol immediately. The identified specimens were stored at −20 °C in the Center of Deep-Sea Research, Institute of Oceanology, CAS, China (sample code: DD201608012FZ, DD201608013FZ, DD201608014FZ, DD201608015FZ) before laboratory works. DNA were extracted using Omega plant DNA mini kit (Omega, D3485-01) following the manufacturer’s protocol. Then, the pair-end library of 350 bp fragments was prepared and sequenced on the Illumina X10 platform in Novogene biotechnology corporation (Tianjin, China). The mitochondrial genomes were assembled using CLC genomic workbench v11 (Qiangen) and reassembled with the extracted raw reads using metawrap pipelines. Annotations were made in the mitos2 (http://mitos2.bioinf.uni-leipzig.de), followed the manual optimization in the seqbuilder in DNAstar v8. The three and one partial mito-genomes have been yielded and deposited in the GenBank (MK343720-MK343723).
Results and discussion
Like other Chaetognath species, three complete mitochondrial genomes we got were small (Marletaz et al. Citation2015; Shen et al. Citation2016; Wei et al. Citation2016, Citation2019), which were 11,610 bp, 11,604 bp, and 12,123 bp in length for Z. bedoti, Z. pulchra, and A. regularis, respectively, due to the absence of 23 (ATP6 and ATP8, and 21 transfer RNA gene) of 37 typical mitochondrial genome coding genes. Considering the gene loss was common phenomenon in Chaetognaths’ mitochondrial genomes (Shen et al. Citation2016), we believed that the gene loss should be an ancestor trait. All obtained mitogenomes were AT preferred (67.5% for Z. bedoti, 65.3% for Z. pulhra, 58.5% for A. regularis, and 59.0% for F. robusta), as found in other invertebrate species. Except for a gene cluster containing ND5, ND4L, ND4, CYTB, and ND6, all other genes located on the positive strand. The largest non-coding regions were all located between ND5 and ND4 genes. Al PCGs start with ATA, ATT, and ATG and terminated either with TAA or with TAG (ND1 and ND5). All tRNA genes possess the typical cloverleaf secondary structures.
The gene orders of the four Chaetognath species were obviously different from those of typical metazoans. Like other invertebrates, gene rearrangements were not special events in the arrow worms (Daniel et al. Citation2004; Faure and Casanova Citation2006; Shen et al. Citation2016). Since the presence and the compositions of transfer RNAs differed in the Chaetognath mito-genomes (Barthélémy and Seligmann Citation2016), we take PCGs and rRNAs into consideration for the further analysis only. As reported before, even species from the same family Spadellidae, the gene order can be remarkably variable (Helfenbein et al. Citation2004; Faure and Casanova Citation2006). Within the family Sagittidae, large-scale rearrangement was also observed in Parasagitta elegans (). Even the previously proposed conserved gene block COX2-srRNA was reshuffled in Parasagitta elegans. The hotspot of gene order rearrangement in Sagittidae is ND4-ND6-ND5-ND4L-CYTB. Three types of rearrangements have been found in five out of ten Sagittidae species within the block, including type 1 in F. robusta, A. regularis and F. refox, type 2 in F. enflata, and type 3 in P. elegans (). Comparing to the ancestral gene order, translocation of ND6 occurred in all scenarios and can be explained by the duplication-random loss model as suggested by Miyamoto (Miyamoto et al. Citation2010). For the third type, strand reverse of ND4 occurred.
Figure 1. (A) Gene arrangements of mitochondrial genomes in 12 Chaetognatha species. Only coding regions are annotated. Genes shown above the bold line in each species are transcribed left to right and those below are transcribed from right to left. Genes below the bold line are reversely transcribed. Dark and light blue indicate fragments of protein genes, yellow and red indicate tRNA and rRNA, respectively. Accession number are listed as below: Parasagitta setosa (KP899760), Parasagitta elegans Parasagitta elegans (KP899784), Flacclsagitta enflata (NC_013814), Zonosagitta nagae (NC_013810), Zonosagitta bedoti (MK343722), Zonosagitta pulchra (MK343723), Aidanosagitta regularis (MK343721), Ferosagitta robusta (MK343720), and Ferosagitta ferox (KT818830.1). (B) Preferred Maximum likelihood tree based on the mitochondrial genomes of 12 Chaetognaths species.
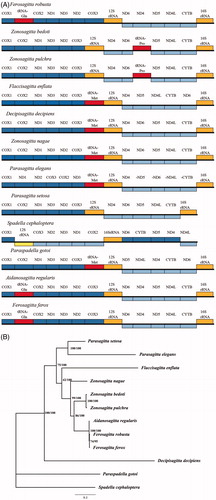
Blastn results based on cox1 sequences confirmed the validity of the species identifications. All the new specimens gave high nucleotide similarity (>97.6) to the corresponding records in GenBank. We investigated the internal phylogenetic relationships of Sagittidae using the amino acids of mitochondrial genome coding genes. Best partitioning scheme was evaluated in Partitionfinder 2.1. Both ML (raxML) and BI (mrbayes) methods were applied to the datasets containing 10 Sagittidae species and two outgroups belonging to Spadellidae. The two methods generated the same tree topology (). Our results questioned the monophyly of Zonosagitta, as the Z. nagae cluster outside the clade composed of Zonosagitta, Aidanosagitta, and Ferosagitta. Monophyly of the Parasagitta and Ferosagitta were supported. In agreement with the large-scaled gene order rearrangement, deep divergence was observed in the Parasagitta, suggesting early speciation or accelerated mutation rates. The Decipisagitta clustered at the basal position of Sagittidae monophylic clade, suggesting its early divergence. The clustering of Flaccisagitta + Aidanosagitta + Ferosagitta revealed in the previous work based on ribosomal RNAs was confirmed in our mitochondrial results (Papillon et al. Citation2006; Gasmi et al. Citation2014).
Geolocation information
Specimen of Zonosagitta bedoti, Zonosagitta pulchra, Aidanosagitta regularis, and Ferosagitta robusta were sampled from Station 1 (122.420°E, 27.051°N), Station 2 (121.074°E, 26.014°N), Station 3(122.030°E, 26.470°N), and Station 4 (120.775°E, 26.343°N), respectively.
Acknowledgements
We are grateful to Ji Peng, technician of IOCAS, for his help in collecting the zooplankton samples.
Disclosure statement
The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Ball EE, Miller DJ. 2006. Phylogeny: the continuing classificatory conundrum of Chaetognaths. Curr Biol. 16:R593–R596.
- Barthélémy RM, Seligmann H. 2016. Cryptic tRNAs in Chaetognath mitochondrial genomes. Comput Biol Chem. 62:119–132.
- Bieri R. 1991. Six new genera in the Chaetognath family Sagittidae. Gulf Res Rep. 8:221–225.
- Daniel P, Yvan P, Xavier C, Yannick LP. 2004. Identification of Chaetognaths as protostomes is supported by the analysis of their mitochondrial genome. Mol Biol Evol. 21:2122–2129.
- Faure E, Casanova JP. 2006. Comparison of Chaetognath mitochondrial genomes and phylogenetical implications. Mitochondrion. 6:258–262.
- Gasmi S, Nve G, Pech N, Tekaya S, Gilles A, Perez Y. 2014. Evolutionary history of Chaetognatha inferred from molecular and morphological data: a case study for body plan simplification. Front Zool. 11:84.
- Giesecke R, González HE, Bathmann U. 2010. The role of the Chaetognath Sagitta gazellae in the vertical carbon flux of the Southern Ocean. Polar Biology. 33:293–304.
- Halanych KM. 1996. Testing hypotheses of Chaetognath origins: long branches revealed by 18S ribosomal DNA. Syst Biol. 45:223–246.
- Helfenbein KG, Fourcade HM, Vanjani RG, Boore JL. 2004. The mitochondrial genome of Paraspadella gotoi is highly reduced and reveals that Chaetognaths are a sister group to protostomes. Proc Natl Acad Sci USA. 101:10639–10643.
- Marletaz F, Parco YL, Liu S, Peijnenburg K. 2015. Extreme mitogenomic variation without cryptic speciation in Chaetognaths. bioRxiv 025957.
- Marlétaz F, Parco YL, Liu S, Peijnenburg KT. 2017. Extreme mitogenomic variation in natural populations of Chaetognaths. Genome Biol Evol. 9:1374–1384.
- Miyamoto H, Machida RJ, Nishida S. 2010. Complete mitochondrial genome sequences of the three pelagic Chaetognaths Sagitta nagae, Sagitta decipiens and Sagitta enflata. Comp Biochem Physiol D. 5:65–72.
- Papillon D, Perez Y, Caubit X, Parco YL. 2006. Systematics of Chaetognatha under the light of molecular data, using duplicated ribosomal 18S DNA sequences. Mol Phylogenet Evol. 38:621–634.
- Shen X, Sun S, Zhao FQ, Zhang GT, Tian M, Ling MT, Wang JF, Chu KH. 2016. Phylomitogenomic analyses strongly support the sister relationship of the Chaetognatha and Protostomia. Zool Scripta. 45:145–233.
- Sun S, Huo Y, Yang B. 2010. Zooplankton functional groups on the continental shelf of the yellow sea. Deep-Sea Res II. 57:1006–1016.
- Thuesen EV, Numachi K, Nemoto T. 1993. Genetic variation in the planktonic Chaetognaths Parasagitta elegans and Eukrohnia hamata. Marine Ecol Prog. 101:243–251.
- Wei S, Li P, Yang M, Zhou L, Yu Y, Ni S, Wang Z, Qin Q. 2016. The mitochondrial genome of the pelagic Chaetognath, Pterosagitta draco. Mitochondrial DNA B. 1:515–516.
- Wei S, Yang M, Dong Y, Qin Q. 2019. Genetic variation analysis of the cosmopolitan Chaetognath Sagitta enflata in the northern South China Sea based on mitochondrial COI gene sequences. Mitochondrial DNA B. 4:5–7.
- Yvan P, Müller G, Harzsch S. 2014. The Chaetognatha: an anarchistic taxon between Protostomia and Deuterostomia. Berlin: De Gruyter. (Deep Metazoan Phylogeny: The Backbone of the Tree of Life: New Insights from Analyses of Molecules, Morphology, and Theory of Data Analysis.
