Abstract
The complete carnation (Dianthus caryophyllus L.) chloroplast (cp) genome was determined and analyzed in this study. The circular cp genome had a total length of 149,595 bp, made up of a pair of 24,808 bp inverted repeats (IRa,b) separated by a large single-copy region (LSC) of 82,883 bp and a small single-copy region (SSC) of 17,096 bp. The overall GC content of the genome is 36.32%. The annotated genome contained 112 unique genes, including 78 protein-coding genes, four ribosomal RNA genes, and 30 tRNA genes. Among these, 16 are duplicated in the inverted repeat regions, and 17 genes contain introns. With the new complete cp genome of carnation, we were also able to correct improperly assembled cp genomes from published data that had indicated a missing one rpl2 copy in the IR regions.
Chloroplasts are double membrane organelles encoding genes essential for photosynthesis and other biochemical processes (Wu et al. Citation2015). This organelle possesses its own single circular DNA chromosome, which is highly conserved across plants containing two inverted repeat copies (IRs) separated by a large single copy region (LSC) and a small single copy region (SSC) (Jansen et al. Citation2005). Published cp genomes provide a valuable genetic resource for types of biological research including plant systematics, DNA barcoding and transformation.
Caryophyllaceae is one of the most diverse and largest family in eudicots (The Angiosperm Phylogeny Group Citation2009). The Dianthus genus consists of nearly 300 species, and the carnation (Dianthus caryophyllus) is well-known due to its use as a cut flower. Given the importance of the carnation to the floricultural industry, it is now grown worldwide and has undergone many rounds of artificial selection and manipulation in the production of new varieties. The genome of carnation had been published (Yagi et al. Citation2014), it would be helpful in understanding and exploring the genetic bases of this species. The cp genome of D. caryophyllus (147,604 bp, MG989277) (Chen et al. Citation2018) was found to be nearly 2kb shorter when compared it with D. longicalyx genome (149,539 bp, KM668208) (Kang et al. Citation2017). Additionally, we found differences in gene content when comparing the two cp genomes. In order to access these discrepancies and provide an improved cp genome for carnation, we reassembled the genome from the previously published genome (Yagi et al. Citation2014) with the original plants saved at NARO Institute of Floricultural Science.
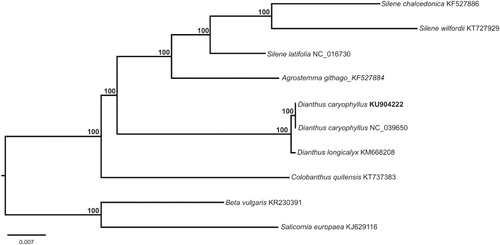
The trimmed and high-quality paired-end reads (DRR014087) from the Yagi et al. (Citation2014) genome assembly were used to complete cp genome using the CLC workbench (ver. 7.01, http://www.clcbio.com) and Ray (ver. 2.3.1, http://denovoassembler.sourceforge.net/). Both reference mapping and de novo assembly methods were employed to assemble the cp genome using Dianthus longicalyx (KM668208) as reference. The final completed cp genome of carnation was targeted as the reference to map back the genome data and check the mapping depth consistency across the whole genome. The final structural features and genes in the cp genome were annotated following the method from Wu and Ge (Wu and Ge Citation2016). The total cp genome length of the reassembled carnation cp genome (KU904222) was 149,595 bp, with LSC at 82,883 bp in length, the SSC at 17,096 bp, and the two IRs (IRa and IRb) at 24,808 bp each. The overall GC content of the cp genome was 36.32% (LSC, 34.00%; SSC, 29.78%; IRs, 42.45%) and contained 112 unique genes, including 78 protein-coding, four rRNA, and 30 tRNA genes. A total of 16 genes were duplicated in the IR regions including seven tRNA, four rRNA, and five protein-coding genes. Among these genes, 17 genes contained introns as 14 of which contained a single intron and three of which contained two introns. The complete ycf1 gene was included in the IR at the SSC/IRa junction while the partial ycf1 gene was pseudogenized and located at IRb/SSC junction. When we compared the newly assembled cp genome (KU904222) with the previously published carnation cp genome (MG989277), we found that the old cp genome (147,604 bp, MG989277) was over 2 kb shorter than the newly assembled genome. This discrepancy in length was caused by the omission of one copy of the rpl2 gene in an IRa region. Additionally, two tRNA genes were incorrectly annotated as trnG-GCC and trnI-CAU. The corrected genome sequence will provide updated information for future comparative cp genome and genetic studies in carnation. To confirm the phylogenetic position of D. caryophyllus, 10 representative species of Caryophyllales were aligned using MAFFT v.7 (https://mafft.cbrc.jp/alignment/server/) and neighbour-joining (NJ) analysis was conducted using MEGA X (https://www.megasoftware.net/). The NJ tree showed that genus Dianthus clustered as one well-supported clade (). The corrected cp genome of D. caryophyllus provides an improved genetic resource for carnation marker-assisted breeding programs.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen S, Xu Z, Zhao Y, Zhong X, Li C, Yang G. 2018. Structural characteristic and phylogenetic analysis of the complete chloroplast genome of Dianthus caryophyllus. Mitochondrial DNA B. 3:1131–1132.
- Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, et al. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. In: Zimmer E, Roalson E, editors. Molecular evolution: producing the biochemical data.Vol. 395. Waltham (MA): Academic Press; p. 348–384.
- Kang JS, Lee BY, Kwak M. 2017. The complete chloroplast genome sequences of Lychnis wilfordii and Silene capitata and comparative analyses with other Caryophyllaceae genomes. PLoS One. 12:e0172924.
- The Angiosperm Phylogeny Group. 2009. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121.
- Wu Z, Ge S. 2016. The whole chloroplast genome of wild rice (Oryza australiensis). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1062–1063.
- Wu Z, Tembrock LR, Ge S. 2015. Are differences in genomic data sets due to true biological variants or errors in genome assembly: an example from two chloroplast genomes. PLoS One. 10:e0118019.
- Yagi M, Kosugi S, Hirakawa H, Ohmiya A, Tanase K, Harada T, Kishimoto K, Nakayama M, Ichimura K, Onozaki T, et al. 2014. Sequence analysis of the genome of carnation (Dianthus caryophyllus L.). DNA Res. 21:231–241.