Abstract
Opisthopappus taihangensis (Ling) Shih is a monotypic species (Anthemideae, Asteraceae) and an endemic endangered plant of China. In this study, we sequenced the complete chloroplast genome of O. taihangensis using Illumina HiSeq X Ten platform. The total length of O. taihangensis chloroplast genome is 151,089 bp, including a large single-copy region of length 82,877 bp, a small single-copy region of length 18304 bp, and a pair of 24,954-bp inverted repeat regions. The chloroplast genome of O. taihangensis has 132 genes, including 87 protein-coding, eight ribosomal RNA, and 37 transfer RNA genes. The overall GC content of the whole genome was 37.5%. The phylogenetic analysis revealed a close relationship between O. taihangensis and Chrysanthemum boreale.
Opisthopappus taihangensis is a monotypic species in the genus and is endemic to China, distributed only in the Taihang Mountain along the borders between Shanxi, Hebei, and Henan Provinces (Shih Citation1979). The sesquiterpene lactones in O. taihangensis have a tonifying effect on heart, as well as anti-cancer, anthelmintic, and analgesic effects; thus, the plant has a high medicinal value (Zeng et al. Citation2010). In this study, for the first time, we sequenced the complete chloroplast genome of O. taihangensis.
The voucher specimen of O. taihangensis were collected from Jiyuan, Henan Province, China (N35°24′15.3″ E112°63′61.8″) and stored at Herbarium of Zhengzhou University. The total DNA was isolated from approximately 0.05 g of dried leaves using the modified CTAB method. The extracted DNA was then purified using the Wizard DNA clean-up system (Promega Corp.). The complete chloroplast genome was sequenced at Novogene Biotech Co. (Beijing, China) using the Illumina HiSeq X Ten platform with a 150-bp shotgun library. Approximately 9.9 GB of 150-bp paired-end reads of O. taihangensis were generated. The obtained raw data, containing some low-quality short sequences, were processed and assembled as described by Bakker et al. (Citation2016) and Wei et al. (Citation2017) (Bakker et al. Citation2016; Wei et al. Citation2017). The sequencing results were mapped to the reference chloroplast genome sequence (Artemisia gmelinii. NCBI accession number NC_031399) using Geneious v 10.1.3 (https://www.geneious.com) software. The complete chloroplast genome was annotated using DOGMA (http://dogma.ccbb.utexas.edu/) and Geneious v 10.1.3 and checked manually. The circular chloroplast genome map was drawn in OGDRAW (Lohse et al. Citation2007). The complete chloroplast genome of O. taihangensis (NCBI accession number MK552323) is 151,089 bp long and has a typical quadripartite structure, with one 82,877-bp long LSC region, one 18,304-bp long SSC region, and two 24,954-bp long IR regions. It contained 132 genes including 86 protein-coding, 37 tRNA, and eight rRNA genes. In addition, six protein-coding, seven tRNA, and four rRNA genes are duplicated in the IR regions. Among annotated genes, eight protein-coding (rps12, rps16, rpl2, rpoC1, ndhA, ndhB, petB, petD, and atpF) and six tRNA (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) genes contained one intron, while two genes (clpP and ycf3) contain two introns. The overall GC content of the chloroplast genome is 37.5%.
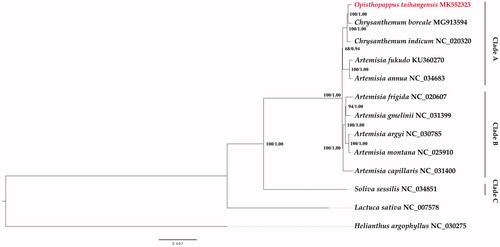
Twelve complete chloroplast genome sequences were downloaded from the NCBI Organelle Genome Resources database. The phylogenetic tree was constructed using RAxML-HPC2 (8.2.10) on XSEDE and MrBayes (3.2.6) on XSEDE, from the CIPRES Science Gateway (Miller et al. Citation2011). Helianthus argophyllus (NC_030275) was set as outgroup. The results showed that Opisthopappus, Soliva, Chrysanthemum, and Artemisia formed one clade (Bootstrap = 100%, Posterior Probability = 1). The phylogenetic relationships of these four genera were consistent with their taxonomic positions that they all belong to the tribe Anthemideae. The Anthemideae Clade were composed of three Clade A, B and C. In Clade A, O. taihangensis and Chrysanthemum species formed a subclade (BS = 100%, PP = 1). Furthermore, O. taihangensis was closer to Chrysanthemum boreale than to Chrysanthemum indicum ().
Figure 1. Phylogenetic tree of 13 species based on the complete chloroplast genome by the maximum likelihood (ML) and Bayesian inference (BI) methods. Helianthus argophyllus was the outgroup. The topology showed in this figure is from the ML tree. The numbers above the branches represent the ML bootstrap values/BI posterior probabilities.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bakker FT, Lei D, Yu JY, Mohammadin S, Wei Z, van de Kerke S, Gravendeel B, Nieuwenhuis M, Staats M, Alquezar-Planas DE, et al. 2016. Herbarium genomics: plastome sequence assembly from a range of herbarium specimens using an Iterative Organelle Genome Assembly pipeline. Biol J Linn Soc. 117:33–43.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Miller MA, Pfeiffer W, Schwartz T. 2011. The CIPRES science gateway: a community resource for phylogenetic analyses. Extreme Digital Discovery; 18–21 July 2011; Salt Lake City, UT, USA.
- Shih C. 1979. Opisthopappus Shih–a new genus of Compositae from China. Acta Phytotaxon Sin. 3:110–112.
- Wei Z, Zhu SX, Van den Berg RG, Bakker FT, Schranz ME. 2017. Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons. Genet Resour Crop Ev. 64:55–71.
- Zeng XY, Luo DL, Liu SN, Liu JX. 2010. Recent advance and future trend of inulin research and development. China Food Addit. 4:222–227.
