Abstract
The complete mitogenomes of one (M-)ale (North America), one Hermaphroditic (Europe), and two (F-)emale (North America and Europe) individuals of the freshwater pearl mussel Margaritifera margaritifera were sequenced. The M-type and F-type (Female and Hermaphroditic) mitogenomes have 17,421 and 16,122 nucleotides, respectively. All with the same content: 13 protein-coding genes, 22 transfer RNA, two ribosomal RNA genes, and one sex-related ORF. The M-type is highly divergent (37.6% uncorrected p-distance) from the F-type mitogenomes. North American and European F-type mitogenomes exhibit low genetic divergence (68 nt substitutions), and the Female and Hermaphroditic European mitogenomes are almost identical, and matching sex-related ORFs.
The Margaritiferidae (Bivalvia: Unionida), comprising 16 extant species, represents the most threatened freshwater mussel family (Lopes-Lima et al. Citation2018). Within this family, the freshwater pearl mussel Margaritifera margaritifera (Linnaeus, 1758) is one of the most threatened species; it is subject to numerous conservation projects and is listed as Endangered globally (Geist Citation2010; Moorkens et al. Citation2018). Margaritifera margaritifera is a long lived species (reaching over 100 years) that generally inhabits cool oligotrophic running waters throughout freshwater systems of northwest Europe and northeast North America (Geist Citation2010; Lopes-Lima, Sousa, et al. Citation2017). As noted for other margaritiferid bivalves, M. margaritifera shows an unusual mitochondrial inheritance process called doubly uniparental inheritance (DUI). Under DUI, both males and females inherit F-type mitochondrial DNA from their mothers, while males also inherit M-type mitochondrial DNA from their fathers, which predominates in gonad tissue (Hoeh et al. Citation1996; Amaro et al. Citation2017). Furthermore, many hermaphroditic species of freshwater mussels seem to have lost DUI and do not possess the M-type mitochondrial genome in their gonad tissues (Breton et al. Citation2011).
The M-type and F-type mitochondrial lineages show high levels of divergence within species of Unionida freshwater mussels and even distinct gene order arrangements (Fonseca et al. Citation2016; Froufe et al. Citation2016; Guerra et al. Citation2017). The Margaritiferidae also exhibit a unique M-type and F-type gene order (Lopes-Lima, Fonseca, et al. Citation2017). Available phylogenetic studies within the family are based on only a few markers still lacking a more robust multi-marker approach (Lopes-Lima et al. Citation2018).
Given this background, the aims of this study are to (1) obtain the whole mitogenomes of male, female, and hermaphroditic specimens of M. margaritifera from North America and Europe; (2) determine and compare the gene order and content of those mitogenomes; and (3) produce phylogenetic analyses using all available F-types and M-type mitogenomes of the Margaritiferidae family.
Four complete mitogenomes of M. margaritifera were sequenced: one M-type and one F-type from a North American male specimen (River Annapolis near Auburn, Canada: 45.014999, -64.856344) and two F-type from European specimens, one from a female (River Ulla near Barazon, Galicia, Spain: approximate coordinates 42.846676, -8.025244) and another from a hermaphrodite (River Tuela near Vinhais, northeast Portugal: approximate coordinates 41.862414, -6.931596). DNA samples are stored at the CIIMAR Institute Unionoid DNA and Tissue Databank (Voucher numbers P2, MM63, 155G, and 165G). Sex was determined for all specimens under a microscope following Hinzmann et al. (Citation2013).
DNA was sheared to ∼500 bp using an M220 Covaris Ultrasonicator (Covaris, Woburn, MA, USA) and processed with the NEBultra Illumina library preparation kit (NEB, Ipswich, MA, USA). Sequencing was performed on the MiSeq (Illumina, San Diego, CA, USA) located at Monash University Malaysia using a run configuration of 2 × 250 bp. Mitogenomes were assembled from the paired-end reads and annotated using an established pipeline (Gan et al. Citation2014). The four mitogenomes have been deposited in the GenBank database under the accession numbers (MK421959 and MK421956; M-type and F-type, respectively for the North American specimens), and (MK421957 and MK421958; for the Spanish and Portuguese F-type European specimens). Sequence divergence (uncorrected p-distance) was assessed using MEGA7 software (Kumar et al. Citation2016). The length of both mitogenome types (M-type: 17,421 nt; and F-type: 16,122 nt) of M. margaritifera sequenced in this study is within the expected range for each gender-specific haplotypes within Margaritiferidae (Guerra et al. Citation2017; Lopes-Lima et al. Citation2017). The larger size of the M-type genomes is expected given the larger cox2 gene and the presence of M-specific coding regions (Breton et al. Citation2009). Both haplotypes have the same gene content: 13 protein-coding genes (PCGs), 22 transfer RNA (trn) and two ribosomal RNA (rrn) genes. ORFs specific to each type of mtDNA, F-orf in the F mitogenome and M-orfs in the M, are also present. Regarding the gene orientation, again, both have the same genes (four PCGs, 20 tRNAs, and two rRNAs) encoded on the heavy strand and the remaining (nine PCGs and two tRNAs) encoded on the complementary strand. The exception is the sex related ORFs, with the M-orf on the complementary strand and the F-orf on the heavy strand, located at different positions. A nucleotide alignment of the mitochondrial genomes shows that the M-type mitogenome is highly divergent (37.6% uncorrected p-distance) from the F-type mitogenomes. The F-type mitogenomes from North America and Europe exhibit a low genetic divergence (68 nt substitutions = 0.04% uncorrected p-distance), with the European mitogenomes of the female and hermaphroditic individuals being almost identical with only 5 nt substitutions. This pattern may reflect a recent (Pleistocene) dispersal event of freshwater pearl mussels from Europe to North America or slow mtDNA substitution rates in this species (Lopes-Lima et al. Citation2018; Zanatta et al. Citation2018). The F-orfs of the European hermaphroditic and female individuals are identical. Secondarily hermaphroditic species generally contain a distinct and longer F-like ORF (Breton et al. Citation2011). Therefore, these results seem to indicate that hermaphroditic individuals of typically dioecious species may maintain their F-type ORFs unchanged.
For the phylogenies, additional mitogenome sequences (M-type and F-type) available from all Margaritiferidae and three Unionidae species were downloaded from GenBank. Each gene sequence was aligned using GUIDANCE v. 1.5 (Penn et al. Citation2010) with the MAFFT v. 7.304 multiple sequence alignment algorithm (Katoh and Standley Citation2013). To build single gene alignments the following GUIDANCE parameters were used: score algorithm: GUIDANCE; bootstraps replicates: 100; Sequence cut-off score: 0.0 (no sequences removed); Column cut-off score: below 0.8. The final concatenated data set included the 13 mitochondrial PCG and the 2 rrn genes for each mitogenome reaching a total length of 13,505 nt. Phylogenetic relationships were estimated by Bayesian inference using MrBayes v. 3.2.6 (Ronquist et al. Citation2012) and Maximum Likelihood using RAxML v. 8.2.10 (Stamatakis Citation2014) HPC Black Box with 100 rapid bootstrap replicates and 20 ML searches at the San Diego Supercomputer Center through the CIPRES Science Gateway (https://www.phylo.org). The final alignment was partitioned in 11 subsets according to the best scheme determined using PartitionFinder v.2.1.1 (Lanfear et al. Citation2016). For the ML a unique GTR model was applied for each partition with corrections for gamma distribution. For the BI, the GTR + G, GTR + I+G, HKY + G, HKY + I+G models were used. Each chain started with a randomly generated tree and ran for 1 × 106 generations with a sampling frequency of 1 tree for every 100 generations. The resultant trees, after discarding the first 25% as burn-in, were combined in a 50% majority rule consensus tree. The final trees were rooted at the split between Male and Female haplotypes (based on previous studies, e.g. Huang et al. Citation2013).
The best obtained phylogenetic BI and ML trees revealed an identical topology (). Both the F and M clades are divided into the two Unionida families, Margaritiferidae, and Unionidae. Maximum support values were obtained for all nodes with two exceptions for the relationships of Pseudunio marocanus both in the female and in the male clades (). The phylogenies are consistent with the systematic divisions of the Margaritiferidae in four genera (Margaritifera, Cumberlandia, Pseudunio, and Gibbosula) and two subfamilies (Margaritiferinae (Margaritifera + Cumberlandia + Pseudunio) and Gibbosulinae (Gibbosula)) (Lopes-Lima et al. Citation2018). The newly sequenced M. margaritifera genomes cluster inside the Margaritifera genus in the F-type clade, being the M-type mitogenome sequence the first available for this genus, following the most recent systematics for the family (Lopes-Lima et al. Citation2018).
Figure 1. Margaritiferidae and Unionidae Bayesian phylogenetic tree of Male and Female mitogenomes sequences based on concatenated nucleotide sequences of 13 mitochondrial protein-coding genes and the two rRNA genes. GenBank accession numbers are behind species names, numbers at the nodes indicate the percentage posterior probabilities and bootstrap support values. The * above the branches indicate posterior probabilities and bootstrap support values > 95%.
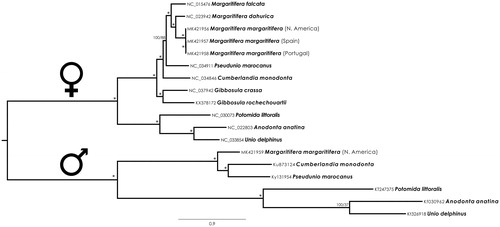
The present results confirm the usefulness of the mitogenomes gene arrangements as diagnostic character for the Margaritiferidae and provide additional confirmation for the systematics of the family as recently proposed by Lopes-Lima et al. (Citation2018). These results also highlight the low intraspecific genetic divergence of M. margaritifera even between specimens from the edges of distribution. Furthermore, the current study provides novel information about mtDNA structure and sequence of hermaphroditic individuals of typical dioecious species providing opportunities for further studies on the sex determination mechanism and mtDNA evolution of freshwater bivalves.
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Amaro R, Bouza C, Pardo BG, Castro J, San Miguel E, Villalba A, Lois S, Outeiro A, Ondina P. 2017. Identification of novel gender-associated mitochondrial haplotypes in Margaritifera margaritifera (Linnaeus, 1758). Zool J Linn Soc. 179:738–750.
- Breton S, Beaupré HD, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, Blier PU, Hoeh WR. 2009. Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionoida) with doubly uniparental inheritance of mtDNA: gender-specific open reading frames and putative origins of replication. Genetics. 183:1575–1589.
- Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol. 28:1645–1659.
- Fonseca MM, Lopes-Lima M, Eackles MS, King TL, Froufe E. 2016. The female and male mitochondrial genomes of Unio delphinus and the phylogeny of freshwater mussels (Bivalvia: Unionida). Mitochondrial DNA B. 1:954–957.
- Froufe E, Gan HM, Lee YP, Carneiro J, Varandas S, Teixeira A, Zieritz A, Sousa R, Lopes-Lima M. 2016. The male and female complete mitochondrial genome sequences of the Endangered freshwater mussel Potomida littoralis (Cuvier, 1798) (Bivalvia: Unionidae). Mitochondrial DNA A. 27:3571–3572.
- Gan HM, Schultz MB, Austin CM. 2014. Integrated shotgun sequencing and bioinformatics pipeline allows ultra-fast mitogenome recovery and confirms substantial gene rearrangements in Australian freshwater crayfishes. BMC Evol Biol. 14:19.
- Geist J. 2010. Strategies for the conservation of endangered freshwater pearl mussels (Margaritifera margaritifera L.): a synthesis of conservation genetics and ecology. Hydrobiologia. 644:69–88.
- Guerra D, Plazzi F, Stewart DT, Bogan AE, Hoeh WR, Breton S. 2017. Evolution of sex-dependent mtDNA transmission in freshwater mussels (Bivalvia: Unionida). Sci Rep. 7:1551.
- Hinzmann M, Lopes -lima M, Teixeira A, Varandas S, Sousa R, Lopes A, Froufe E, Machado J. 2013. Reproductive cycle and strategy of Anodonta anatina (L., 1758): Notes on hermaphroditism. J Exp Zool A Ecol Genet Physiol. 319:378–390.
- Hoeh WR, Stewart DT, Sutherland GW, Zouros E. 1996. Multiple origins of gender-associated mitochondrial DNA lineages in bivalves (Mollusca: Bivalvia). Evol. 50:2276–2286.
- Huang XC, Rong J, Liu Y, Zhang MH, Wan Y, Ouyang S, Zhou CH, Wu XP. 2013. The complete maternally and paternally inherited mitochondrial genomes of the endangered freshwater mussel Solenaia carinatus (Bivalvia: Unionidae) and implications for Unionidae taxonomy. PLoS One. 8:e84352
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Lopes-Lima M, Bolotov IN, Do VT, Aldridge DC, Fonseca MM, Gan HM, Gofarov MY, Kondakov AV, Prié V, Sousa R, et al. 2018. Expansion and systematics redefinition of the most threatened freshwater mussel family, the Margaritiferidae. Mol Phylogenetics Evol. 127:98–118.
- Lopes-Lima M, Sousa R, Geist J, Aldridge DC, Araujo R, Bergengren J, Bespalaya Y, Bódis E, Burlakova L, et al. 2017. Conservation status of freshwater mussels in Europe: state of the art and future challenges. Biol Rev. 92:572–607.
- Lopes-Lima M, Fonseca MM, Aldridge DC, Bogan AE, Gan HM, Ghamizi M, Sousa R, Teixeira A, Varandas S, Zanatta D, et al. 2017. The first Margaritiferidae male (M-type) mitogenome: mitochondrial gene order as a potential character for determining higher-order phylogeny within Unionida (Bivalvia). J Molluscan Stud. 83:249–252.
- Moorkens E, Cordeiro J, Seddon MB, von Proschwitz T, Woolnough D. 2018. Margaritifera margaritifera (errata version published in 2018). The IUCN Red List of Threatened Species 2018: e.T12799A128686456.
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38:W23–W28.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Zanatta DT, Stoeckle BC, Inoue K, Paquet A, Martel AL, Kuehn R, Geist J. 2018. High genetic diversity and low differentiation in North American Margaritifera margaritifera (Bivalvia: Unionida: Margaritiferidae). Biol J Linn Soc. 123:850–863.
