Abstract
The complete mitochondrial genome was determined for Arctic staghorn sculpin Gymnocanthus tricuspis (Reinhardt, 1830), which is considered an important part of Arctic food webs. The mitogenome sequence is 16,570 bp in length containing 2 ribosomal RNA genes, 22 tRNA genes, 13 protein-coding genes, and 2 non-coding regions consist of control region (CR) and light-strand replication origin (OL). The overall base composition of the mitogenome was estimated to be A 23.2%, T 29.0%, C 28.8%, and G 19.0%. The complete mitochondrial genome data will provide useful genetic information for future genetic variation identification and genetic diversity evaluation of this important species in Arctic Ocean.
The sculpin family Cottidae comprises 264 species of benthic scorpaeniform fishes inhabiting cold, northern coastal waters from shallow depths to about 300 m (Mecklenburg et al. Citation2018). Of these species, the Arctic staghorn sculpin Gymnocanthus tricuspis (Reinhardt 1830) is one of most abundant and widespread Arctic sculpins and is considered an important part of Arctic food webs with little about mitogenome of this species (Smith et al. Citation1997; Mecklenburg et al. Citation2011; Gray et al. Citation2016). Thus, we present the complete mitochondrial genome of G. tricuspis, which will be an important genetic resource to assist in phylogenetic analysis.
The sample of G. tricuspis was collected from the Chukchi Sea (168.850°W, 69.603°N) during the eighth Chinese Arctic Research Expedition and deposited in the Third Institute of Oceanography, Ministry of Natural Resources of China. The complete G. tricuspis mitogenome was amplified using a long-PCR technique, and then subsequent sequencing was accomplished by primer walking method.
The complete mitogenome (mitogenome) sequence of G. tricuspis (GenBank accession no. MK204282) is 16,570 bp in length containing 2 ribosomal RNA (12S and 16S) genes, 22 transfer RNA genes (tRNA), 13 protein-coding genes (PCGs), and 2 non-coding regions consist of control region (CR) and light-strand replication origin (OL). Except for ND6 and eight tRNAs (-Gln, -Ala, -Asn, -Cys, -Tyr, -Ser, -Glu, and -Pro), all other genes are encoded on the heavy strand. The overall base composition of the mitogenome was estimated to be A 23.2%, T 29.0%, C 28.8%, and G 19.0%. All PCGs but COI (GTG) use ATG as the start codon. Three PCGs are ended with incomplete stop codons (COII, ND4, Cyt b), and the other genes are ended with TAG (ND1, ND3, ND6) and TAA (ND2, CO I, ATPase8, ATPase6, CO III, ND4L, ND5).
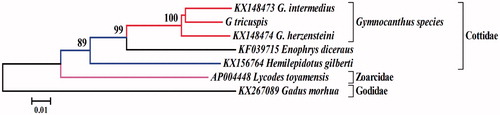
Phylogenetic relationship was constructed using NJ algorithm based on complete mitogenome sequence of G. tricuspis and other 6 Arctic fish species. MEGA version 6.06 (Tamura et al. Citation2013) was used to construct neighbour-joining (NJ) tree based on Kimura 2-parameter (K2P) model. The neighbour-joining tree () showed that they were obviously differentiated between G. tricuspis and other species. Gymnocanthus tricuspis first clustered together with Gymnocanthus intermedius and Gymnocanthus herzensteini and formed a monophyly in the genus Gymnocanthus, and then they constituted a sister-group relationship with the other genus species E. diceraus, which belonged to the same family Cottide.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Gray BP, Norcross BL, Beaudreau AH, Blanchard AL, Seitz AC. 2016. Food habits of Arctic staghorn sculpin (Gymnocanthus tricuspis) and shorthorn sculpin (Myoxocephalus scorpius) in the northeastern Chukchi and western Beaufort Seas. Deep-Sea Res II. 135:111–123.
- Mecklenburg CW, Moller PW, Steinke D. 2011. Biodiversity of Arctic marine fishes: taxonomy and zoogeography. Mar Biodiv. 41:109–140.
- Mecklenburg CW, Lynghammar A, Johannesen E, Byrkjedal I, Christiansen JS, Dolgov AV, Karamushko OV, Mecklenburg TA, Møller PR, Steinke D, Wienerroither RM. 2018. Marine fishes of the arctic region. Conservation of arctic flora and fauna. Akureyri, Iceland: Arctic Council Secretariat. ISBN: 978-9935-431-69-1.
- Smith RL, Barber WE, Vallerino M, Gillispie J, Ritchie A. 1997. Population biology of the Arctic Staghorn Sculpin in the Northeastern Chukchi Sea. Am Fish Soc Symp. 19:133–139.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.