Abstract
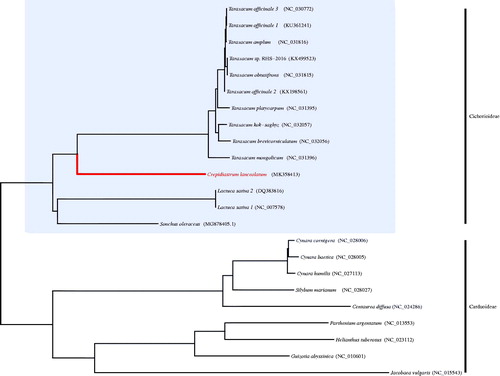
The characteristic of complete chloroplast (cp) genome sequence of Crepidiastrum lanceolatum, a member of the subfamily Cichorioideae with unresolved phylogenetic relationship among the related taxa, was firstly reported in this study. It was 152,748 bp and contained a large-single copy region (SSC) (84,022 bp) and a small-single copy region (LSC) (18,568 bp) which were separated by two inverted repeat (IR) regions (25,079 bp). In total, 133 genes were identified and they were consisted of 86 coding genes, 8 rRNA genes, 37 tRNA genes, and 2 pseudogenes. Molecular phylogenetic analysis showed that the genus Crepidiastrum was a sister group of the genus Taraxacum in subfamily Cichorioideae.
Although the occurrence frequency of Asteraceae in the world, the lack of a well-resolved backbone phylogeny for the family caused the problem to understand their evolutionary history. Recently, 9 subfamilies and 25 tribes system of the family were suggested based on the phylogenomics approach (Mandel et al. Citation2017). Using the complete chloroplast genome data has been regarded as a powerful tool for building accurate and reliable phylogenetic tree. Genus Crepidiastrum, which are distributed in the Eastern Asia, is one of the difficult taxa to define the circumscription and relationship among the related genus like Paraixeris, Youngia, Crepis, and Ixeridium (Peng et al. Citation2014) because of their similarity in morphology. For resolving this issue, we tried to obtain the complete chloroplast (CP) genome sequence of this genus and expect to get massive data for genetic approach.
We collected the plant material from Busan of Korea and the voucher specimen was deposited at the Herbarium of Kyungpook National University (KNU). cp genome of Crepidiastrum lanceolatum (MK358413) was sequenced by HiSeq4000 of Illumina. Totally 40,906,620 paired-end reads (2 × 151 bp) were obtained and 5,407,032 reads were assembled to the reference chloroplast genomes after reads end trimming with an error probability limit of 0.01. And then assembled reads were de novo assembled using the Geneious assembler. Using the assembled contigs, we conducted to align and repeat the procedure up to make a single contig. cp genome was annotated using Geneious version 10.2.3 (Biomatters, Ltd.; Kearse et al. Citation2012) with manual correction and tRNAScan-SE (Lowe and Eddy Citation1997) for tRNA gene. The average coverage of this cp genome was 5130. The phylogenetic tree was constructed with related Asteraceae members based on the concatenated 78 coding genes using RAxML (Stamatakis Citation2014).
It was typical circular form with 152,748 bp in length and comprised a large-single copy region (LSC, 84,022 bp), a small-single copy region (SSC, 18,568 bp), and two inverted repeat regions (IR, 25,079 bp). It was composed of 133 genes and they were identified 86 coding genes, 8 rRNA genes, 37 tRNA genes, and 2 pseudogenes. Two pseudogenes were ycf1 and trnH positioned in the boundary area of IR. In the phylogenetic tree, it was clear that the genus Crepidiastrum was a sister group of the genus Taraxacum in subfamily Cichorioideae (). Comparing to Taraxacum chloroplast genomes, it was approximately 1300 bp longer due to the length variation at IR between tow genera. For comprehensively understand the relationship among the genus Crepidiastrum and allied taxa, we will to expand the plant sample and analyse the complete chloroplast genome sequences of them in the near future.
Disclosure statement
The authors have no conflicts of interest in this study.
Additional information
Funding
References
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Mandel JR, Barker MS, Bayer RJ, Dikow RB, Gao TG, Jones KE, Keeley S, Kilian N, Ma H, Siniscalchi CM, et al. 2017. The Compositae tree of life in the age of phylogenomics. J Syst Evol. 55:405–410.
- Peng YL, Zhang Y, Gao XF, Tong LJ, Li L, Li RY, Zhu ZM, Xian JR. 2014. Aphylogenetic analysis and new delimitation of Crepidiastrum (Asteraceae, tribe Cichorieae). Phytotaxa. 159:241–255.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.