Abstract
Calocedrus is a genus of the family Cupressaceae. Among this genus, C. decurrens is the only species native to western North America. In this study, the complete chloroplast genome (plastome) of C. decurrens was determined using Illumina sequencing platform. The plastome lacking one copy of inverted repeat (IR) was 127,112 bp in length. The plastome contained 118 unique genes, including 83 protein-coding genes (PCGs), 31 transfer RNA genes (tRNAs), and 4 ribosomal RNA genes (rRNAs). Nine PCGs and six tRNAs contained one intron each, two PCGs contained two introns each. The overall GC content of the plastome was 34.8%. Phylogenomic analysis of 24 representative plastome within the Cupressaceae family revealed that C. decurrens was sister to the remaining Calocedrus species, and C. rupestris was sister to the clade comprising of C. macrolepis and C. formosana.
Calocedrus is a genus of the family Cupressaceae (Farjon Citation2005). It is native to eastern Asia and western North America. Four extant species are in this genus, i.e. C. macrolepis, C. formosana, C. rupestris, and C. decurrens. But C. decurrens is the only one of the four extant species native to western North America (Farjon Citation2005). It is widely called as incense-cedar in the genus. The tree is an important timber species and is widely grown as a handsome ornamental. The chloroplast and mitochondrial DNA of this species are both paternally inherited (Neale et al. Citation1991). Up to now, the plastomes of C. macrolepis, C. formosana, and C. rupestris have been reported in our previous studies (Qu, Wu, et al. Citation2017; Qu Citation2019). In this study, we sequenced the complete plastome of C. decurrens and determined its phylogenetic placement within Calocedrus, which will contribute to widespread phylogenetic and conservation genetic studies.
Fresh leaves of a single individual of C. decurrens were sampled from Humboldt (California, USA; 40°20′17″N, 123°32′49″W) and voucher specimen (Stoughton 1760) was deposited at Kunming Institute of Botany Chinese Academy of Sciences. Total genomic DNA was extracted by the modified CTAB method described in Wang et al. (Citation2013). Due to limited fresh sample, the plastid DNA plastid DNA was not directly extracted (Liu et al. Citation2017). The total genomic DNA was used for library construction and paired-end (PE) sequencing on the Illumina MiSeq instrument at Kunming Institute of Botany (Kunming, China). In total, 2.4 G raw reads were obtained and assembled using the Organelle Genome Assembler (OGA) pipeline (https://github.com/quxiaojian/OGA). The details of the assembly process are described in Qu (Citation2019): (1) mapping raw reads to the reference plastome of C. macrolepis (Qu, Wu, et al. Citation2017) with Bowtie v2.3.4 (Langmead and Salzberg Citation2012); (2) assembling mapped plastome reads to contigs by Spades v3.7.1 (Bankevich et al. Citation2012); (3) using plastome contigs as seeds to recruit overlapped reads from raw reads, then using recruited overlapped reads as new seeds, and iterating this step until no overlapped reads are recruited by OGA; (4) assembling mapped plus recruited reads to final scaffolds using Spades. (5) connecting the path of plastome scaffolds by Bandage v8.0 (Wick et al. Citation2015). The plastome was annotated by a newly developed software package Plastid Genome Annotator (PGA, https://github.com/quxiaojian/PGA), coupled with manual correction using Geneious v9.1.4 (Kearse et al. Citation2012). Physical map of the circular plastome was drawn by OGDRAW (Lohse et al. Citation2013). The annotated complete plastome has been deposited into the GenBank with the accession number MK241803.
The complete plastome of C. decurrens was 127,112 bp in length. The overall GC content was 34.8%. The plastome of C. decurrens, like other plastomes in cupressophytes, lacks one copy of the canonical inverted repeat (IR) regions (Qu, Wu, et al. Citation2017). This plastome contained a total of 118 unique genes, including 83 protein-coding genes (PCGs), 31 transfer RNA genes (tRNAs) and 4 ribosomal RNA genes (rRNAs). Eleven PCGs contained introns, of which nine PCGs (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, and rps16) contained one intron and two PCGs (rps12 and ycf3) contained two introns.
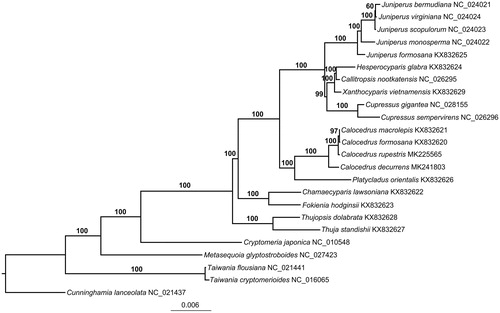
The maximum likelihood (ML) phylogenetic tree was reconstructed by RAxML v8.2.10 (Stamatakis Citation2014), including tree robustness assessment using 1000 rapid bootstrap replicates with the GTRGAMMA substitution model, based on the alignment of 73 shared PCGs (Qu, Jin, et al. Citation2017) using MAFFT (Katoh and Standley Citation2013). The ML phylogenetic tree showed that all Calocedrus species formed a monophyletic clade and the monotypic genus Platycladus (Platycladus orientalis) was sister to Calocedrus (). Within Calocedrus, C. decurrens was firstly diverged and was sister to the remaining Calocedrus species, and C. rupestris was sister to the clade comprising C. macrolepis and C. formosana. This complete plastome will provide valuable information for future population genomic studies, and will establish a useful basis for the utilization of this species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Farjon A. 2005. A monograph of Cupressaceae and Sciadopitys. Edinburgh (UK): Royal Botanic Gardens.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Liu F, Jin Z, Wang Y, Bi YP, Melton JT. 2017. Plastid genome of Dictyopteris divaricata (Dictyotales, Phaeophyceae): understanding the evolution of plastid genomes in brown algae. Mar Biotechnol. 19:627–637.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Neale DB, Marshall KA, Harry DE. 1991. Inheritance of chloroplast and mitochondrial DNA in incense-cedar (Calocedrus decurrens). Can J Forest Res. 21:717–720.
- Qu X-J. 2019. Complete plastome sequence of an endangered species, Calocedrus rupestris (Cupressaceae). Mitochondrial DNA B. 4:762–763.
- Qu X-J, Jin J-J, Chaw S-M, Li D-Z, Yi T-S. 2017. Multiple measures could alleviate long-branch attraction in phylogenomic reconstruction of Cupressoideae (Cupressaceae). Sci Rep. 7:41005.
- Qu X-J, Wu C-S, Chaw S-M, Yi T-S. 2017. Insights into the existence of isomeric plastomes in Cupressoideae (Cupressaceae). Genome Biol Evol. 9:1110–1119.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang HY, Jiang DF, Huang YH, Wang PM, Li T. 2013. Study on the phylogeny of Nephroma helveticum and allied species. Mycotaxon. 125:263–275. English.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.