Abstract
Abies balsamea is native to the mountains of North America and often used as Christmas Trees. In this study, we reported the complete chloroplast genome of A. balsamea. The complete chloroplast genome is 121,574 bp in length, comprising a large single copy (LSC) region of 67,211 bp, a small single copy (SSC) region of 53,835 bp, and two inverted repeat regions (IRa and IRb) of 264 bp each. The genome contains 114 genes and they were 68 peptide-encoding genes, 35 tRNA genes, 4 rRNA genes, 6 open reading frames and one pseudogene. Loss of ndh genes and inverted repeat sequences were identified in this genome. The phylogenetic analysis confirms that the Abies species are supported as monophyletic, with high bootstrap values.
Abies consists of about 50 species, which are widely distributed in the Northern Hemisphere including East Asia, North America and Europe (Liu Citation1971; Farjon Citation2001). Recently, the complete chloroplast genomes of several fir species in East Asia and Europe had been reported (A. beshanzuensis, A. alba) and were found to be highly conserved (Yi et al. Citation2015; Shao et al. Citation2018). However, the complete chloroplast genomes of fir species from North America have never been surveyed. Abies balsamea Mill., a North American fir, is native to most of eastern and central Canada (Newfoundland west to central Alberta) and the northeastern United States (Minnesota east to Maine, and south in the Appalachian Mountains to West Virginia) (Liu Citation1971). It tends to grow in cool climates, ideally with a mean annual temperature of 4 °C and occurs at altitudes of 100–2100 m (Farjon Citation2001).
The plant material was collected from the Royal Botanic Gardens Edinburgh, UK. Specimens were given identification numbers and registered in the herbarium of Institute of Botany, CAS (PE), with voucher no. MWC 36531. The complete chloroplast (cp) genome was sequenced by HiSeq4000 of Illumina, and finally, 10.3 million high-quality clean reads (150 bp PE read length) were obtained. In total, ca. 10.2 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and tRNAscan-SE v1.3.1 (Schattner et al. Citation2005) were used to align, assemble, and annotate the plastome.
The complete chloroplast genome consists of 121,574 bp for Abies balsamea (GenBank: MH706725). The circle genome was comprised of a large single copy (LSC) region of 67,211 bp, a small single copy (SSC) region of 53,835 bp, and two inverted repeat regions (IRa and IRb) of 264 bp each. The GC content is 38.2%, with the SSC region having higher GC content (39.2%) than the LSC (37.3%) and IR (39.0%) regions. The genome contains 114 genes and they were 68 peptide-encoding genes, 35 tRNA genes, 4 rRNA genes, 6 open reading frames, and 1 pseudogene. All ndh genes have been lost in the genome. Short inverted repeat sequences were detected in 52-kb inversion points of the cp genome, which consist of trnS-psaM-ycf12-trnG and trnG-ycf12-psaM-trnS (1183 bp). Interestingly, such missing of ndh genes and inverted repeats had been reported in several members of the genus Abies (A. beshanzuensis, A. koreana) (Yi et al. Citation2015; shao et al. Citation2018).
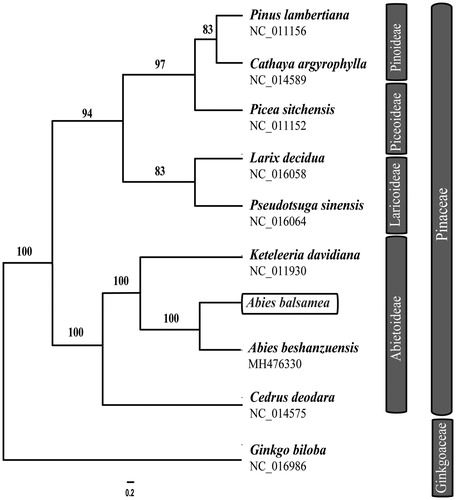
To investigate the phylogenetic position of Abies balsamea, nine chloroplast genomes were selected in Pinaceae with Ginkgo biloba (Ginkgoaceae) as the outgroup. These genomes were fully aligned with MAFFT v7.3 (Suita, Osaka, Japan) (Katoh and Standley Citation2013), and the maximum likelihood (ML) inference was performed using GTRþIþC model with RAxML v.8.2.1 (Karlsruhe, Germany) (Stamatakis Citation2014) on the CIPRES cluster service (Miller et al. Citation2010). The ML tree revealed that A. balsamea and A. beshanzuensis formed a monophyletic group with bootstrap support values of 100% ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Farjon A. 2001. World checklist and bibliography of conifers, 2nd edn. London, UK: The Royal Botanic Gardens, Kew.
- Liu TS. 1971. A monograph of the genus Abies. Taiwan: National Taiwan University.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of 2010 Gateway Computing Environments Workshop (GCE 2010); November 14; New Orleans, LA.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Shao YZ, Hu JT, Fan PZ, Liu YY, Wang YH. 2018. The complete chloroplast genome sequence of Abies beshanzuensis, a highly endangered fir species from south China. Mitochondrial DNA B. 3:923–924.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Yi DK, Yang JC, So S, Joo MJ, Kim DK, Shin CH, Lee YM, Choi K. 2015. The complete plastid genome sequence of Abies koreana (Pinaceae: Abietoideae). Mitochondrial DNA. 27:1.