Abstract
The complete mitochondrial genome (mitogenome) of Minois dryas (Lepidoptera: Nymphalidae: Satyrinae) is determined to be 15,195 bp in length, including 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 non-coding AT-rich region. The mitogenome includes a cox1 gene with an atypical CGA(R) start codon and five genes (cox1, cox2, nad5, nad4, nad1) exhibiting incomplete stop codons. The 385-bp AT-rich region contains the motif ATAGA followed by a 19-bp poly-T stretch and a microsatellite-like (TA)6 element preceded by the ATTTA motif. The phylogenetic analysis indicated that M. dryas is formed a monophyletic group with other Satyrinae species.
The subfamily Satyrinae, comprising approximately described species, is one of the most diverse groups of butterflies (Peña and Wahlberg Citation2008), comprising over a third of the Nymphalidae diversity and found on all continents except Antarctica (Ackery et al. Citation1999). Owing to their species richness and ecological diversification, taxonomy, and phylogeny, Satyrinae are still standing as a controversial issue, and need to be elucidated (Marín et al. Citation2011; Yang and Zhang Citation2015). The insect mitochondrial genome (mitogenome) is highly useful in phylogenetic relationships analysis, due to their unique features (Salvato et al. Citation2008; Wu et al. Citation2014; Timmermans et al. Citation2016).
Here, the complete mitogenome of Minois dryas (Satyrinae: Satyrini) is revealed based on the samples collected from Minshan District in Gansu Province, China (coordinates: E104° 03′, N34° 43′), and kept in the laboratory at −20 °C under the accession number SQH-20130722. Total genomic DNA was extracted from thorax muscle of an adult individual using the Sangon Animal Genome DNA Extraction Kit (Shanghai, China). The raw sequences were assembled and annotated using the BioEdit 7.0 (Hall Citation1999) and MEGA7.0 software (Kumar et al. Citation2016) with reference to the mitogenome of Davidina armandi (GenBank accession NC_028505).
The complete mitogenome of M. dryas (GenBank accession is MK521433) is 15,195 bp in length, containing typical insect 13 protein-coding genes (PCGs), two ribosomal RNA genes (rRNAs) and 22 transfer RNA genes (tRNAs), along with an AT-rich control region. Its gene arrangement and orientation are consistent with those of other known nymphalid mitogenomes (e.g. Wu et al. Citation2014; Shi et al. Citation2018; Lalonde and Marcus Citation2019). The nucleotide composition is also biased toward AT nucleotides (80.2%).
All PCGs are initiated by typical ATN, with the exception of the cox1 which uses the unusual CGA(R) as observed in other sequenced nymphalid mitogenomes (e.g. Kim et al. Citation2010; Shi et al. Citation2015). The mitogenome contains five PCGs (cox1, cox2, nad5, nad4, nad1) with single-nucleotide (T) stop codons completed by adding 3’ A residues post-transcriptionally. All tRNAs harbor the typical cloverleaf structures commonly found in insects, except for trnS1(AGN), whose dihydrouridine (DHU) arm is replaced by a simple loop. The rrnL and rrnS genes are 1339 bp and 772 bp in length, with the A + T contents of 84.5% and 85.7%, respectively. The putative AT-rich region is 385 bp long (94.0% A + T content) with several structures characteristic of lepidopteran, such as the 19-bp poly-T stretch preceded by the ATAGA motif neighboring the rrnS gene, and a microsatellite-like (AT)6 element preceded by the ATTTA motif.
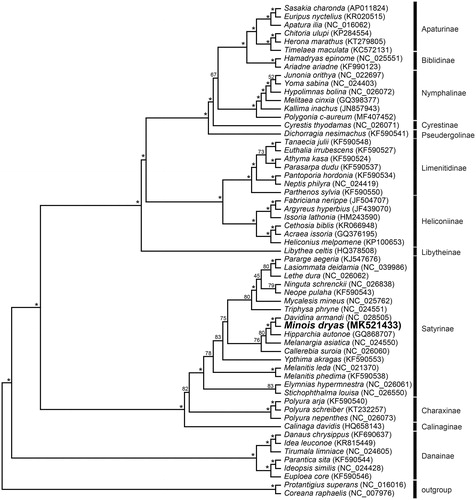
We performed a maximum-likelihood (ML) phylogenetic analysis with RAxML (version 7.2.6) using nucleotide sequences of 13 PCGs and 2 rRNAs to understand the phylogenetic relationship of M. dryas with other nymphalids (see for details). The phylogenetic analysis indicated that M. dryas formed a monophyletic group with other Satyrinae species, which was recovered as the sister group to Charaxinae, same as results from other recent analyses (Wu et al. Citation2014; Shi et al. Citation2015).
Figure 1. The maximum likelihood (ML) phylogenetic tree of Minois dryas and other nymphalid species. Phylogenetic reconstruction was done from a concatenated matrix of 13 protein-coding mitochondrial genes and 2 ribosomal RNA genes regions in the mitochondrial genome. The numbers beside the nodes are percentages of 1000 bootstrap values (* ≥ 85%). Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
Additional information
Funding
References
- Ackery PR, de Jong R, Vane-Wright RI. 1999. The butterflies: Hedyloidea, Hesperoidea, and Papilionoidea. Berlin: de Gruyter; p. 263–300.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Kim MJ, Wan X, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lalonde MML, Marcus JM. 2019. The complete mitochondrial genome of the Madagascar banded commodore butterfly Precis andremiaja (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 4:277–279.
- Marín MA, Peña C, Freitas A, Wahlberg N, Uribe SI. 2011. From the phylogeny of the Satyrinae butterflies to the systematics of Euptychiina (Lepidoptera: Nymphalidae): history, progress and prospects. Neotrop Entomology. 40:1–13.
- Peña C, Wahlberg N. 2008. Prehistorical climate change increased diversification of a group of butterflies. Biol Lett. 4:274.
- Salvato P, Simonato M, Battisti A, Negrisolo E. 2008. The complete mitochondrial genome of the bag-shelter moth Ochrogaster lunifer (Lepidoptera, Notodontidae). BMC Genomics. 9:331.
- Shi QH, Sun XY, Wang YL, Hao JS, Yang Q. 2015. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS ONE. 10:e0124349.
- Shi QH, Xing JH, Liu XH, Hao JS. 2018. The complete mitochondrial genome of comma, polygonia c-aureum (Lepidoptera: Nymphalidae: Nymphalinae). Mitochondrial DNA Part B. 3:53–55.
- Timmermans M, Lees DC, Thompson MJ, Sáfián S, Brattström O. 2016. Mitogenomics of ‘Old World Acraea’ butterflies reveals a highly divergent ‘Bematistes’. Mol Phylogenet Evol. 97:233–241.
- Wu LW, Lin LH, Lees DC, Hsu YF. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics. 15:468.
- Yang MS, Zhang YL. 2015. Phylogenetic utility of ribosomal genes for reconstructing the phylogeny of five Chinese satyrine tribes (Lepidoptera: Nymphalidae). Zookeys. 488:105–120.
