Abstract
Aiolocaria hexaspilota (Hope 1831) is a specialized predator of leaf beetles (Chrysomelinae). We have determined the first mitochondrial genome of A. hexaspilota. The circular mitogenome of A. hexaspilota consists of 17,549 bp including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNAs, and a single large non-coding region of 1,603 bp. The base composition was AT-biased (80.1%). The control region of this mitochondrial genome was the shortest among the five complete mitochondrial genomes in Conccinellidae. Phylogenetic trees strongly supported the monophyletic relationship of Coccinellinae. A. hexaspilota mitochondrial genome will be a fundamental resource for understanding the molecular phylogenetic relationship of a species-rich family Coccinellidae of Coleoptera.
Aiolocaria hexaspilota (Hope 1831), one of the large ladybirds up to 13 mm in size, belongs to the family Coccinellidae of Coleoptera. It distributes in Asian countries including Korea, Japan, southern China, Taiwan, the Himalayas, and northern India (Yoneya and Takabayashi Citation2013). Larvae and adults of A. hexaspilota prey on eggs and larvae of several leaf beetles in Chrysomelinae including Gastrolina depressa, major pest of walnut trees (Matsura Citation1976; Chang and Park Citation2011), and Plagiodera versicolora, leaf feeder of Salicaceous plants (Nakamura et al. Citation2005; Yoneya et al. Citation2009; Yoneya et al. Citation2012).
We completed the mitochondrial genome of A. hexaspilota from DNA extracted using CTAB-based DNA extraction method manually (iNtRON Biotechnology, Inc., Gyounggi province, Korea). Samples were captured at Ganghwa-gun, Incheon, Republic of Korea (37°43′36″N, 126°24′52″E) in 2018. DNA sample was deposited in National Institute of Agricultural Sciences of Rural Development Administration. Sequencing was conducted by HiSeqX (Macrogen Inc., Seoul, Korea) with filtering by Trimmomatic 0.33 (Bolger et al. Citation2014) and de novo assembled was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and MITObim (Hahn et al. Citation2013). Post processes including confirmation of the assembled sequence were conducted by SOAPGapCloser 1.12 (Zhao et al. 2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate mitochondrial genome based on the partial mitochondrial genome of Propylea japonica CL131 (KM244660; Tang et al. Citation2014) with ARWEN (Laslett and Canbäck Citation2008) for annotating tRNAs.
Aiolocaria hexaspilota mitochondrial genome length is 17,549 bp (GenBank Accession number: MK583344) containing 13 protein-coding genes, 2 rRNAs, and 22 tRNAs. The nucleotide composition is AT-biased (A + T ratio is 80.1%). The size of tRNAs ranges from 55 bp to 70 bp are similar to those of other partial or complete Coccinellidae mitochondrial genomeswhich ranges from 54 bp to 79 bp. Gene order of partial or complete Coccinellidae mitochondrial genomes is identical. The control region, presumably corresponding to the single largest non-coding AT-rich region (1,603 bp, A + T ratio is 84.3%), is the shortest among five complete mitochondrial genomes in Conccinellidae (Kim et al. Citation2012; Behere et al. Citation2016).
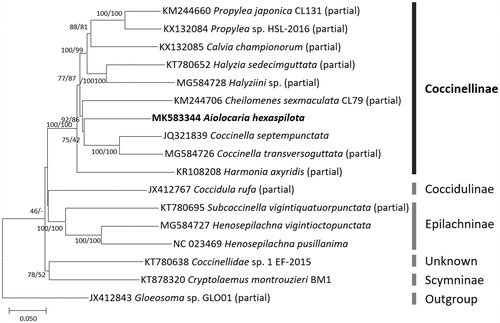
Phylogenetic relationship was inferred from 17 complete or partial mitochondrial genome sequences, 16 in Coccinellidae, and one in Corylophidae. Multiple sequence alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) and unaligned areas including rRNAs and AT-rich regions due to partial mitochondrial genomes were trimmed. Bootstrapped neighbor joining and maximum likelihood trees were constructed using MEGA X (Kumar et al. Citation2018). A. hexaspilota was clustered with two Coccinella species in the phylogenetic trees (). In addition, monophyletic relationship among species of subfamily Coccinellinae was inferred, which agrees with previous phylogeny studies (Seago et al. Citation2011; Escalona et al. Citation2017). Coccinellidae consisting of over 6,000 species in 360 genera presents high biodiversity in Coleoptera (Escalona et al. Citation2017); however, only a few genera have been known about phylogenetic relationship with several partial nuclear and mitochondrial genes (Seago et al. Citation2011; Escalona et al. Citation2017; Wang et al. Citation2018). Therefore, A. hexaspilota whole mitochondrial genome will be a fundamental resource for understanding the phylogeny and evolution of Coccinellidae.
Figure 1. Neighbor-joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 16 Coccinellidae and one Corylophidae complete or partial mitochondrial genomes: Aiolocaria hexaspilota (MK583344 in this study), Coccinella septempunctata (JQ321839), Coccidula rufa (JX412767; partial genome), Propylea japonica isolate CL131 (KM244660; partial genome), Cheilomenes sexmaculata isolate CL79 (KM244706), Harmonia axyridis (KR108208; partial genome), Coccinellidae sp. 1 EF-2015 (KT780638), Halyzia sedecimguttata (KT780652; partial genome), Subcoccinella vigintiquatuorpunctata (KT780695; partial genome), Cryptolaemus montrouzieri isolate BM1 (KT878320; partial genome), Propylea sp. HSL-2016 (KX132084; partial genome), Calvia championorum (KX132085; partial genome), Coccinella transversoguttata (MG584726; partial genome), Henosepilachna vigintioctopunctata (MG584727), Halyziini sp. HA (MG584728; partial genome), Henosepilachna pusillanima (NC_023469), and Gloeosoma sp. GLO01 (JX412843) as an outgroup species. Right bars present subfamilies' name in family Coccinellidae. Phylogenetic tree was drawn based on neighbor joining phylogenetic tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Behere G, Firake D, Tay W, Azad Thakur N, Ngachan S. 2016. Complete mitochondrial genome sequence of a phytophagous ladybird beetle, Henosepilachna pusillanima (Mulsant)(Coleoptera: Coccinellidae). Mitochondrial DNA A. 27:291–292.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Chang S-J, Park I-K. 2011. Morphological and ecological study of Gastrolina depressa Baly (Coleoptera: Chrysomelidae). Korean J Appl Entomol. 50:253–256.
- Escalona HE, Zwick A, Li H-S, Li J, Wang X, Pang H, Hartley D, Jermiin LS, Nedvěd O, Misof B, et al. 2017. Molecular phylogeny reveals food plasticity in the evolution of true ladybird beetles (Coleoptera: Coccinellidae: Coccinellini). BMC Evol Biol. 17:151.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Rese. 41:e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim MJ, Wan X, Kim I. 2012. Complete mitochondrial genome of the seven-spotted lady beetle, Coccinella septempunctata (Coleoptera: Coccinellidae). Mitochondrial DNA. 23:179–181.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Matsura T. 1976. Ecological studies of a coccinellid, Aiolocaria hexaspilota Hope. I. Interaction between field populations of A. hexaspilota and its prey, the walnut leaf beetle (Gastrolina depressa Baly). Jpn J Ecol. 26:147–156.
- Nakamura M, Utsumi S, Miki T, Ohgushi T. 2005. Flood initiates bottom‐up cascades in a tri‐trophic system: host plant regrowth increases densities of a leaf beetle and its predators. J Anim Ecol. 74:683–691.
- Seago AE, Giorgi JA, Li J, Slipiński A. 2011. Phylogeny, classification and evolution of ladybird beetles (Coleoptera: Coccinellidae) based on simultaneous analysis of molecular and morphological data. Mol Phylogenet Evolut. 60:137–151.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, Zhou X. 2014. Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166–e166.
- Wang Z-L, Wang T-Z, Zhu H-F, Wang Z-Y, Yu X-P. 2018. DNA barcoding evaluation and implications for phylogenetic relationships in ladybird beetles (Coleoptera: Coccinellidae). Mitochondrial DNA A. 42:e166.
- Yoneya K, Kugimiya S, Takabayashi J. 2009. Can herbivore‐induced plant volatiles inform predatory insect about the most suitable stage of its prey? Physiol Entomol. 34:379–386.
- Yoneya K, Takabayashi J. 2013. Interaction–information networks mediated by plant volatiles: a case study on willow trees. J Plant Interact. 8:197–202.
- Yoneya K, Uefune M, Takabayashi J. 2012. An apparent trade-off between direct and signal-based induced indirect defence against herbivores in willow trees. PLoS One. 7:e51505.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics.12 (Suppl 14):S2.
