Abstract
The complete mitochondrial genome of Dasypogon diadema (Insecta: Diptera) was sequenced using the Illumina MiSeq` platform. Its mt-genome spans over 16,947 bp with a GC content of 26.6% containing 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes. The phylogenetic relationship of Dasypogon diadema and 11 other dipteran species was reconstructed and the phylogenetic position confirmed.
Keywords:
Dasypogon diadema resembles one of the largest European robber fly species that occurs in central and southern Europe. It is known to prey on well defensive and venomous insects, especially hymenopterans (Geller-Grimm Citation1995; Poulton Citation2009). Robber flies (Asilidae) are the only known family of Dipterans in which both genders employ venom for their predatory lifestyle (Kahan Citation1964; von Reumont et al. Citation2014; Drukewitz et al. Citation2018). Recently, it was shown that members of the Asilidae secrete toxins which are similar to known neurotoxins from venoms of cone snails, spiders, scorpions and assassin bugs (Drukewitz et al. Citation2018). Here, the first complete mitochondrial genome of a robber fly is assembled. An Illumina library was prepared using extracted DNA from muscle tissue of a single female specimen collected near Milas (France) (42°42′00.3″N 2°40′31.3″E). The whole DNA was sequenced on a MiSeq platform and a voucher of the specimen stored at the Justus Liebig University Gießen. All reads were trimmed afterward with Trimmomatic (a minimum length of 70pb, a minimum Phred score of 30) and the quality was checked in FastQC (Andrews; Bolger et al. Citation2014). The mitochondrial genome was assembled using MITObim (Hahn et al. Citation2013) with the cytochrome c oxidase subunit 1 gene as a starting seed. The resulting sequence is 16947 bp long and is composed of A: 39.9%; C: 16.3%; G: 10.3% and T: 33.5%.
Genome annotation was conducted on the MITOS webserver (Bernt et al. Citation2013) and the results were visualized in Geneious v9. If necessary, the annotations were corrected manually. Compared to other dipteran mitochondrial genomes (Jiang et al. Citation2016; Qi et al. Citation2017) the arrangement of the protein-coding genes in Dasypogon diadema is identical.
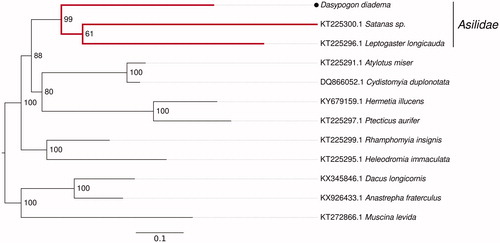
The phylogenetic position of Dasypogon diadema was tested performing a maximum-likelihood analysis of its 13 protein-coding mitochondrial genes in RAxML v.8.2.0 (Stamatakis Citation2014) For the phylogenetic tree reconstruction, suitable dipteran mitochondrial sequences were mined in GenBank. All translated protein-coding mitochondrial genes were separately aligned with MAFFT E-INS-I (Katoh and Standley Citation2013). After concatenating the single genes to one alignment using a custom python script, the RAxML analyses were conducted with the model MTART + GAMMA + I and 1000 Bootstraps.
Dasypogon diadema (MK061306) groups together with the robber fly sequences obtained from GeneBank (KT225300.1; KT225296.1), which confirms its phylogenetic position within asilids in the dipteran clade.
The first complete mitochondrial genome of a robber fly provides the basis for further studies to reveal the still unclear phylogeny of asilid dipterans.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Andrews S. Babraham Bioinformatics – FastQC A Quality Control tool for High Throughput Sequence Data. [accessed 2018 Aug 1]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Drukewitz SH, Fuhrmann N, Undheim EAB, Blanke A, Giribaldi J, Mary R, Laconde G, Dutertre S, von Reumont BM. 2018. A dipteran’s novel sucker punch: evolution of arthropod atypical venom with a neurotoxic component in robber flies (asilidae, diptera). Toxins (Basel). 10.
- Geller-Grimm F. 1995. Autökologische Studien an Raubfliegen (Diptera: Asilidae) auf Binnendünen des Oberrheintalgrabens.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Jiang F, Pan X, Li X, Yu Y, Zhang J, Jiang H, Dou L, Zhu S. 2016. The first complete mitochondrial genome of Dacus longicornis (Diptera: Tephritidae) using next-generation sequencing and mitochondrial genome phylogeny of Dacini tribe. Sci Rep. 6:36426.
- Kahan D. 1964. The toxic effect of the bite and the proteolytic activity of the saliva and stomach contents of the robber flies (Diptera Asilidae.). Isr J Zool. 13:47–57.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Poulton EB. 2009. XVI. Predaceous insects and their prey. Trans R Entomol Soc Lond. 54:323–410.
- Qi Y, Xu J, Tian X, Bai Y, Gu X. 2017. The complete mitochondrial genome of Hermetia illucens (Diptera: Stratiomyidae). Mitochondrial DNA B. 2:189–190.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- von Reumont B, Campbell L, Jenner R. 2014. Quo Vadis Venomics? A roadmap to neglected venomous invertebrates. Toxins (Basel). 6:3488–3551.