Abstract
The mitochondrial genome of Bengalia sp. is documented in the present study, making it the first representative of the subfamily Bengaliinae with nearly complete mitogenome sequenced. This 15,748 bp mitogenome consists of 22 transfer RNA genes, 13 protein-coding genes, 2 ribosomal RNA genes, and 1 noncoding control region. The genome organization, base composition, and codon usage of the mitogenome are noted. Phylogenetic analysis strongly supported that Bengaliinae is the sister group to the clade (Chrysomyinae, (Calliphorinae, Lucillinae). This study portrayed the systematic position of Bengaliinae within the Calliphoridae and provided mitogenomic data for further study on the evolution of the Calliporadae.
The Calliphoridae (Diptera: Oestroidea) is one of the most important forensic insect groups, while its monophyly has been controversial (Rognes Citation1997). Bengaliinae is a subfamily of Calliphoridae with many species feeding on a varied range of animals, especially termites (Rognes Citation2011). Here, we reported and characterized the nearly complete mitogenome of Bengalia sp., which make it the first representative of subfamily Bengaliinae for further study of termite parasites and phylogeny of the Calliphoridae.
The adult specimens of Bengalia sp. were collected from Jianfengling National Forest Park, Hainan province, China on 13 June 2016. The specimens were deposited in the Museum of Beijing Forestry University, Beijing, China (Accession number: BFU0876-0877). The genomic DNA was extracted from the middle leg of a dry specimen using the DNeasy Blood and Tissue kit (QIAGEN Sciences, Valencia, CA, USA). The mitogenome was pulled out referring the procedures of Gillett et al. (Citation2014) after sequencing pooled DNA of several calyptrate species using Illumina HiSeq 2500 platform (Illumina, San Diego, CA). The mitogenome was annotated using the MITOS webserver (Bernt et al. Citation2013). The annotated genes were further revised by comparing with other calyptrates using MEGA version6 (Tamura et al. Citation2013).
The mitochondrial genome (GenBank accession number: MK591038) is 15,748 bp in length and contains 22 transfer RNA (tRNA) genes, 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, and 1 noncoding control region, which is similar to other dipteran mitogenomes reported before (e.g. Nelson et al. Citation2012; Zhang et al. Citation2016; Yan et al. Citation2017; Ding et al. Citation2019). The nucleotide composition is biased towards A and T (77%) with 39.3% of A, 37.7% of T, 13.7% of C, and 9.3% of G. Most of the 13 PCGs used ATN as the start codon (ATG for COII, ATP6, COIII, ND4, ND4L and CYTB; ATT for ND2, ND5, and ND6; ATA for ND3 and ND1; ATC for ATP8), except that COI begins with codon TCG. The stop codon TAA is assigned to most of the PCGs (ND2, ATP8, ATP6, COIII, ND3, ND4L, ND6, ND1), whereas incomplete stop codon T is used by four PCGs (COI, COII, ND5, and ND4) and only CYTB stop with the codon TAG.
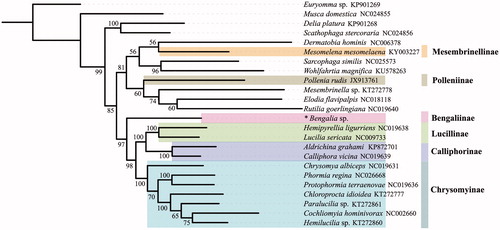
With mitogenomes of 24 calyptrate species, phylogenetic analysis was performed employing IQ-TREE (Nguyen et al. Citation2015) under models automatically assigned for each partition by ModelFinder (Kalyaanamoorthy et al. Citation2017). Euryomma sp. (Fanniidae) was chosen to root the tree. According to the phylogenetic result (), the Calliphoridae are not a monophyletic group, which is congruent with previous studies, as Pollenia and Mesembrinella are nested within other oestroid clades (Kutty et al. Citation2010; Junqueira et al. Citation2016). The monophyly of the Lucillinae, Calliphorinae, and Chrysomyinae is consistently fully supported and clustered as (Chrysomyinae, (Calliphorinae, Lucillinae). The result also show that the Bengaliinae, represented by Bengalia sp., is the sister group to the clade (Chrysomyinae, (Calliphorinae, Lucillinae).
Acknowledgements
The authors are grateful to Mr. Chao Wang for specimens collection and Miss Haoran Sun for specimens identification. We also thank Prof. Hu Li for his help in bioinformatics analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Ding SM, Zhao CJ, Yang D. 2019. Complete mitochondrial genome of Heterostomus Sp. (Diptera, Xylophagidae). Mitochondrial DNA B. 4:884–885.
- Gillett C, Crampton-platt A, Timmermans M, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31:2223–2237.
- Junqueira ACM, Azeredo-Espin AML, Paulo DF, Marinho MAT, Tomsho LP, Drautz-Moses DI, Purbojati RW, Ratan A, Schuster SC. 2016. Large-scale mitogenomics enables insights into Schizophora (Diptera) radiation and population diversity. Sci Rep. 6:21762.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Kutty SN, Pape T, Wiegmann BM, Meier R. 2010. Molecular phylogeny of the Calyptratae (Diptera: Cyclorrhapha) with an emphasis on the superfamily Oestroidea and the position of Mystacinobiidae and McAlpine’s fly. Syst Entomol. 35:614–635.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511:131–142.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Rognes K. 1997. The Calliphoridae (Blowflies) (Diptera: Oestroidea) are not a monophyletic group. Cladistics. 13:27–66.
- Rognes K. 2011. A review of the monophyly and composition of the Bengaliinae with the description of a new genus and species, and new evidence for the presence of Melanomyinae in the Afrotropical Region (Diptera, Calliphoridae). Zootaxa. 2964:1–60.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yan LP, Zhang M, Gao YY, Thomas P, Zhang D. 2017. First mitogenome for the subfamily Miltogramminae. (Diptera: Sarcophagidae) and its phylogenetic implications. Eup J Entomol. 114:442–449.
- Zhang D, Yan LP, Zhang M, Chu HJ, Cao J, Li K, Hu DF, Thomas P. 2016. Phylogenetic inference of calyptrates, with the first mitogenomes for Gasterophilinae (Diptera: Oestridae) and Paramacronychiinae (Diptera: Sarcophagidae). Int J Biol Sci. 12:489–504.