Abstract
Xanthoceras sorbifolium is an important woody oil seed tree in North China. In this study, the complete mitochondrial genome of X. sorbifolium was sequenced using Illumina Hiseq and PacBio sequencing technique. The mitogenome is 575,633 bp in length and the GC content is 45.71%. The genome consists of 42 protein-coding genes, 4 ribosomal-RNA genes, and 24 transfer-RNA genes. Phylogenetic analysis based on protein-coding genes showed that X. sorbifolium was close with the species in Bombacaceae and Malvaceae family.
Yellowhorn (Xanthoceras sorbifolium), a genus of Xanthocera in Sapindaceae family, is an oil-rich seed tree in China, whose seeds contain about 55–66% oil (Bi and Guan Citation2014; Gao et al. Citation2002). Xanthoceras sorbifolium could grow well in barren soil, has great potential for edible oil production in the future (Bi and Guan Citation2014). The complete chloroplast genome has been sequenced in the previous study by Chen and Zhang (Citation2017). In this study, we reported the complete mitogenome of X. sorbifolium using high-throughput sequencing and PacBio sequencing technique. The annotated mitogenome of X. sorbifolium has been deposited into GenBank under accession number MK333231.
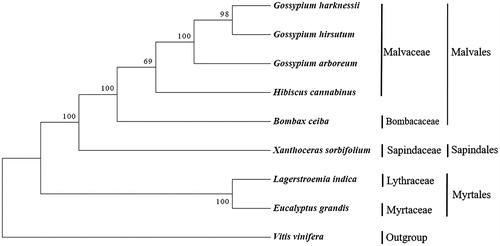
The young leaves were collected from a new variety ‘Zhongshi 4’ in Zhangwu, Liaoning, China, which has more female flowers and higher yield. The voucher specimens were deposited in the State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China. The mt DNA was isolated using an improved extraction method (Chen et al. Citation2011). The mitochondrial genome was reconstructed using a combination of the Pacbio Sequel data and the Illumina Hiseq data. At first, the genome framework was assembled by both Illumina and Pacbio data using SPAdes v3.10.1 (Dmitry et al. Citation2016). Then, verifying the assembly and completing the circle or linear characteristic of the mitochondrial genome and filling gaps if there were any. The last step includes clean reads were mapped to the assembled mitochondrial genome to correct the wrong bases, judge if there were any insertion and deletion. The mitochondrial genes were annotated using homology alignments and de novo prediction. The complete mitogenome of X. sorbifolium is 575,633 bp in length and the GC content was 45.71%. The mitogenome of X. sorbifolium comprises 42 protein-coding genes, 24 tRNA genes, and 4 rRNA genes. The total sequence length of the protein-coding gene was 35,011 bp and the average length of genes was 834 bp. The phylogenetic relationships of X. sorbifolium and eight other plant mitogenome from GenBank were reconstructed using Bayesian analysis based protein-coding genes (). Sequences were aligned using MEGA X software (Kumar et al. Citation2018). Phylogenetic analysis confirmed that X. sorbifolium was close with the species in Bombacaceae and Malvaceae family. All members in the phylogenetic tree provided the relationships among the Vitales, Sapindales, Malvales and Myrtales. The complete mitochondrial genome reported here will provide a useful resource for further exploring the phylogeny, population genetic diversity, and conservation genetics of X. sorbifolium.
Figure 1. The phylogenetic tree based on nine complete mitochondrial genomes. Accession number: Gossypium harknessii (JX536494.1), Gossypium hirsutum (JX065074.1), Gossypium arboretum (NC035073.1), Hibiscus cannabinus (NC035549.1), Bombax ceiba (NC038052.1), Lagerstroemia indica (NC035616.1), Eucalyptus grandis (NC040010.1), Vitis vinifera (NC012119.1), Xanthoceras sorbifolium (MK333231).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bi QX, Guan WB. 2014. Isolation and characterisation of polymorphic genomic SSRs markers for the Endangered tree Xanthoceras sorbifolium Bunge. Conserv Genet Resour. 6:895–898.
- Chen J, Guan R, Chang S, Du T, Zhang H, Xing H. 2011. Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS One. 6:e17662.
- Chen SY, Zhang XZ. 2017. Characterization of the complete chloroplast genome of Xanthoceras sorbifolium, an Endangered oil tree. Conserv Genet Resour. 9:595–598.
- Dmitry A, Anton K, Jeffrey S, Pevzner PA. 2016. HYBRIDSPADES: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32(7):1009–1015.
- Gao SM, Ma K, Du XH, Li FL. 2002. Advances in research on Xanthoceras sorbifolium. Chinese Bull Bot. 19:296–301.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
