Abstract
The complete mitochondrial genome was sequenced from the Antarctic harpacticoid copepod Tigriopus kingsejongensis. The sequenced genome size was 14,940 bp, possessing different gene order and contents to those of the congeneric species T. japonicus and T. californicus in the genus Tigriopus. The mitochondrial genome of T. kingsejongensis has 13 protein-coding genes (PCGs), 2 rRNAs, and 22 tRNAs. Of the 13 PCGs, CO1, ND3, ATP6, and CO3 genes had incomplete stop codons T–, T–, TA–, and T–, respectively. Furthermore, the stop codons of the remaining 11 PCGs were TAA or TAG, while the start codon of 13 PCGs was ATG (CO1, ND4, Cytb, ND2, ND1, CO2, ND6, ATP6, CO3 genes), ATT (ND5, ATP8 genes), and ATA (ND3, ND4L genes), respectively. The nucleotide of 13 PCGs of T. kingsejongensis mitogenome showed 53.42, 52.17, and 49.10% similarities to the copepods T. japonicus, T. californicus, and Amphiascoides atopus, respectively, while the amino acid similarities were 64.38, 63.86, and 62.40%, respectively.
Until now, 13 species are retrieved from the genus Tigriopus (Huys Citation2001). Of them, five complete mitochondrial genomes of two Tigriopus japonicus (Machida et al. Citation2002; Jung et al. Citation2006) and three Tigriopus californicus (Burton et al. Citation2007) have been reported with a high similarity of the gene structure of the mitochondrial genome. However, despite the reported morphometry with CO1 gene (Park et al. Citation2014), RNA-seq (Kim et al. Citation2016), and the whole genome (Kang et al. Citation2017), the taxonomical consideration of the copepod Tigriopus kingsejongensis based on complete mitogenome is still unavailable (Huys Citation2001; Park et al. Citation2014). Since T. kingsejongensis can be a good model for studying environmental adaptation (e.g. ultraviolet ray) in the genus Tigriopus (Han et al. Citation2016; Kang et al. Citation2017), we report the complete mitochondrial genome of the Antarctic copepod T. kingsejongensis to better understand the phylogenetic position of T. kingsejongensis within the genus Tigriopus.
The Antarctic copepod T. kingsejongensis was originally collected at 62°14′10.29″S, 58°46′38.67″W by Dr. Sanghee Kim (Korea Polar Research Institute, South Korea) and maintained in the Department of Biological Science, Sungkyunkwan University in South Korea (kindly provided by Dr. Sanghee Kim). We sequenced 800 bp mate-pair library of T. kingsejongensis from whole body genomic DNA using the Illumina HiSeq 2500 platform (GenomeAnalyzer, Illumina, San Diego, CA). De novo assembly was conducted by Minia (version 3.2.0) (https://github.com/GATB/minia) with K-mer 320. Of the assembled T. kingsejongensis 139,592 contigs with Newbler (version 2.9; identity 100) (http://www.454.com), nine supercontigs were obtained. After a manual curation of nine supercontigs with Consed (version 19.0) (http://www.phrap.org/consed/consed.html), a single contig was mapped to the mitochondrial DNA of T. japonicus.
The total length of the complete mitochondrial genome of T. kingsejongensis was 14,940 bp (GenBank accession no. MK598762). The mitochondrial genome of T. kingsejongensis contained 13 protein-coding genes (PCGs), 2 rRNAs, and 22 tRNAs. The direction of 13 PCGs of T. kingsejongensis was mostly different to those of a congeneric species T. japonicus (Machida et al. Citation2002; Jung et al. Citation2006) and T. californicus (Burton et al. Citation2007). Of the 13 PCGs in T. kingsejongensis, CO1, ND3, ATP6, and CO3 genes had incomplete stop codons T–, T–, TA-, and T–, respectively. Furthermore, the stop codons of the remaining 11 PCGs were TAA or TAG, whereas the start codon of 13 PCGs was ATG (CO1, ND4, Cytb, ND2, ND1, CO2, ND6, ATP6, CO3 genes), ATT (ND5, ATP8 genes), and ATA (ND3, ND4L genes), respectively. Between the two sister species (T. kingsejongensis and T. japonicus), the similarities of amino acids and nucleotides of 13 PCGs were 64.38 and 53.42%, respectively, which demonstrated higher similarity compared to that of T. californicus (63.86% for amino acids and 52.17% for nucleotides) and Amphiascoides atopus (62.40% for amino acids and 49.10% for nucleotides).
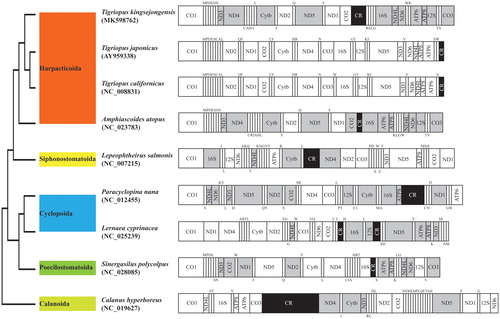
The placement of T. kingsejongensis among four Harpacticoida species with known complete copepod mitogenomes with outgroup (Calanoida Calanus hyperboreus) was shown in . Among the three Tigriopus spp., T. kingsejongensis showed strikingly different patterns of mitogenomic arrangement compared to two congeneric species (T. japonicus and T. californicus), yet was closely situated to T. japonicus compared to T. californicus. This phylogenetic tree of the copepods supports the finding of Wang et al. (Citation2011). This information will be helpful for a better understanding of mitogenome evolution in the genus Tigriopus.
Figure 1. Phylogenetic analysis. We conducted comparison of the mitochondrial genomes of nine copepods. Amino acids of 13 PCGs gene from nine copepods were aligned using MEGA software (ver. 10.0.1) with the ClustalW alignment algorithm. To establish the best-fit substitution model for phylogenetic analysis, the model with the lowest Bayesian Information Criterion (BIC) and Akaike Information Criterion (AIC) scores were estimated using a maximum likelihood (ML) analysis. According to the results of model test, maximum likelihood phylogenetic analyses were performed with the GTR + G+I model. Gray, the opposite direction of PCGs; Black, control regions; Upper and lower single letters indicate tRNA clockwise and anticlockwise, respectively.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Burton RS, Byrne RJ, Rawson PD. 2007. Three divergent mitochondrial genomes from California populations of the copepod Tigriopus californicus. Gene. 403:53–59.
- Huys R. 2013. Copepoda - Harpacticoida. In: Costello, M.J. et al. (eds.) European register of marine species: a check-list of the marine species in Europe and a bibliography of guides to their identification. Collection Patrimoines Naturels 50:268–280.
- Han J, Puthumana J, Lee M-C, Kim S, Lee J-S. 2016. Different susceptibilities of the Antarctic and temperate copepods Tigriopus kingsejongensis and T. japonicus to ultraviolet (UV) radiation. Mar Ecol Prog Ser. 561:99–107.
- Jung S-O, Lee Y-M, Park T-J, Park HG, Hagiwara A, Leung KMY, Dahms H-U, Lee W, Lee J-S. 2006. The complete mitochondrial genome of the intertidal copepod Tigriopus sp. (Copepoda, Harpactidae) from Korea and phylogenetic considerations. J Exp Mar Biol Ecol. 333:251–262.
- Kang S, Ahn DH, Lee JH, Lee SG, Shin SC, Lee J, Min GS, Lee H, Kim HW, Kim S, et al. 2017. The genome of the Antarctic-endemic copepod, Tigriopus kingsejongensis. GigaScience. 6:1–9.
- Kim H-S, Lee B-Y, Han J, Lee YH, Min GS, Kim S, Lee J-S. 2016. De novo assembly and annotation of the Antarctic copepod (Tigriopus kingsejongensis) transcriptome. Mar Genomics. 28:37–39.
- Machida RJ, Miya MU, Nishida M, Nishida S. 2002. Complete mitochondrial DNA sequence of Tigriopus japonicus (Crustacea: Copepoda). Mar Biotechnol. 4:406–417.
- Park E-O, Lee S, Cho M, Yoon SH, Lee Y, Lee W. 2014. A new species of the genus Tigriopus (Copepoda: Harpacticoida: Harpacticidae) from Antarctica. Proc Biol Soc Wash. 127:138–154.
- Wang M, Sun S, Li C, Shen X. 2011. Distinctive mitochondrial genome of Calanoid copepod Calanus sinicus with multiple large non-coding regions and reshuffled gene order: useful molecular markers for phylogenetic and population studies. BMC Genomics. 12:73.
