Abstract
Calanthe delavayi, an endemic terrestrial orchid distributed in southwestern China. In this study, the complete chloroplast genome (cpDNA) sequence of C. delavayi was determined from Illumina pair-end sequencing data. With a total length of 150,181 bp in length and includes two inverted repeat regions (IRs) of 23,721 bp each, which were separated by a large single copy region (LSC) 87,392 bp and a small single copy region (SSC) 18,693 bp. The chloroplast genome contained 135 genes, including protein-coding genes, tRNA genes, and eight rRNA genes. Phylogenetic analysis indicates that C. delavayi, C. davidii, and C. triplicata cluster together, placed them within tribe Collabieae.
Calanthe delavayi Finet is an alpine plant endemic to temperate zones of southwestern China at 2700–3500 m (Perner and Cribb Citation2002; Chen et al. Citation2010). On the basis of its floral morphology, the specie was treated as a member of genera Phaius in Pridgeon et al. (Citation2005), but it was confirmed as a member of genera Calanthe due to molecular evidence by Zhai et al. (Citation2014). Calanthe delavayi has become a key species in the study of taxonomy owing to its complex taxonomic history between Calanthe and Phaius, meanwhile Calanthe (∼216 species) and Phaius (∼45 species) are two biggest genera in tribe Collabieae (Clayton and Cribb Citation2013; Chase et al. Citation2015), at present, there are uncertainties in the classification of tribe Collabieae. To better understand of its evolution history and genetic information, we characterized the complete chloroplast genome sequence of C. delavayi as a resource for future genetic studies.
The total genomic DNA was extracted from dry leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced based on the Illumina pair-end technology. The leaf sample was collected from Huanglong Valley, Huanglong Nature Reserve, Sichuan, China (N 32°45′, E 103°49′) and the DNA stored at the Fujian Agriculture and Forestry University (N0.S42). The clean reads were firstly aligned to C. triplicata (GenBank accession No. KF753635) (Yang et al. Citation2014) and then assembled into contigs in the software CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark). The assembled chloroplast genome was annotated using DOGMA, and the annotation was corrected using Geneious (Kearse et al. Citation2012). The physical map of the new chloroplast genome was generated using OGDRAW (Lohse et al. Citation2013). The accurate new annotated complete chloroplast genome was submitted to GenBank with accession number MK388860. The complete chloroplast genome of C. delavayi is 150,181 base pairs (bp) in length, containing a large single-copy (LSC) region of 83,559 bp, a small single-copy (SSC) region of 16,486 bp, and two inverted repeat (IR) regions of 25,068 bp. The new sequence possesses total 135 genes, of which 114 were unique genes, including 73 protein-coding genes, eight rRNA genes, and 40 tRNA genes. Among all of these genes, eight rRNA genes (i.e. 4.5S, 5S, 16S, and 23S rRNA), eight protein-coding genes (i.e. ndhB, rpl2, rpl23, rps12, rps19, rps7, ycf15, and ycf2), and 10 tRNA genes (i.e. trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnM-CAU, trnN-GUU, trnR-ACG, trnS-UGA, and trnV-GAC) occur in double copies. The overall GC-content of the whole plastome is 36.9%, whereas the corresponding values of the LSC, SSC, and IR regions are 34.5, 29.4, and 43.3%, respectively.
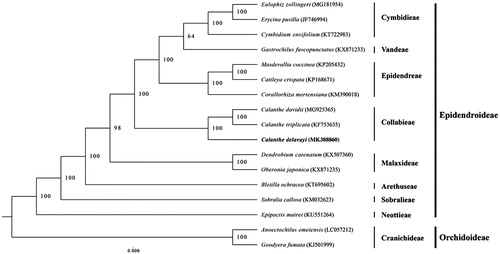
The phylogenetic analysis was carried out with C. delavayi and 16 other complete cp genome of species from Orchidaceae, comprising of 14 Epidendroideae species (Bletilla ochracea, Calanthe triplicata, Calanthe davidii, Cymbidium ensifolium, Erycina pusilla, Eulophia zollingeri, Cattleya crispata, Corallorhiza mertensiana, Masdevallia coccinea, Dendrobium catenatum, Oberonia japonica, Epipactis mairei, Sobralia callosa, and Gastrochilus fuscopunctatus,) and two Orchidoideae species (Anoectochilus emeiensis and Goodyera fumata) as outgroup. The sequences were aligned using HomBlocks pipeline (Bi et al. Citation2018). RAxML-HPC2 on XSEDE version 8.2.10 (Stamatakis Citation2014) was used to construct a maximum likelihood tree, the branch support was computed with 1000 bootstrap replicates. C. delavayi, C. davidii, and C. triplicata cluster together with strong bootstrap support placed them within the tribe Collabieae ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Berg C, Schuiteman A. 2015. An updated classification of Orchidacea. Bot J Linn Soc. 177:151–174.
- Chen S, Liu Z, Zhu G. 2010. Orchidaceae flora of China. vol. 25. Beijing: Science Press, Missouri Botanical Garden Press.
- Clayton D, Cribb P. 2013. The genus Calanthe. Borneo: Natural History Publications.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Perner H, Cribb P. 2002. Orchid wealth. Alp Gard. 70:285–294.
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. 2005. Genera Orchidacearum Volume 4: Epidendroideae. Oxford: OUP.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14:1024–1031.
- Zhai JW, Zhang GQ, Li L, Wang MN, Chen LJ, Chung SW, Rodriguez F, Francisco-Ortega J, Lan SR, Xing FW, Liu ZJ. 2014. A new phylogenetic analysis sheds new light on the relationships in the Calanthe alliance (Orchidaceae) in China. Mol Phylogenet Evol. 77:216.