Abstract
In this study, the complete mitogenome of an entomopathogenic fungus Isaria farinosa ARSEF 3 was assembled and annotated. This circular mitogenome is 24,225 bp in length and consists of two rRNA genes (rnl and rns), 25 tRNA genes, and 14 standard protein-coding genes of the oxidative phosphorylation system. Only one intron is identified. This group IA intron invades rnl and carries an ORF coding for ribosomal protein S3. Phylogenetic analysis based on concatenated mitochondrial nucleotide sequences confirms I. farinosa in Cordycipitaceae although the taxonomy of strains deposited as I. farinosa in culture collections awaits reappraisal.
Isaria farinosa, formerly Paecilomyces farinosus, is the type species of Isaria and the anamorph of Cordyceps memorabilis (Pacioni and Frizzi Citation1978). It occurs on seven insect orders (Coleoptera, Diptera, Hemiptera, Homoptera, Hymenoptera, Lepidoptera, and Thysanoptera) and also on spiders (Araneae) (Zimmermann Citation2008). Genomes of two strains of the fungus, KACC47486 (LUFE01000000) and MTCC4114 (JMNC01000000), have been sequenced, but its mitogenome remains unknown. Herein, we present the mitogenome of I. farinosa ARSEF 3, which is isolated from an eastern tent caterpillar in Chapin Beach, Dennis, Massachusetts, USA. The strain is obtained from USDA-ARS Collection of Entomopathogenic Fungal Cultures (ARSEF) and is identical to ATCC 24319.
Total DNA of ARSEF 3 was randomly sheared to fragments of 400–500 bp, followed by sequencing on an Illumina HiSeq 4000 platform in 2 × 150 bp reads. Mitogenome was de novo assembled from clean reads using MITObim (Hahn et al. Citation2013) and then annotated as described previously (Zhang et al. Citation2017).
The mitogenome of ARSEF 3 (GenBank accession: MK512381) is a circular molecule of 24,225 bp with an AT content of 73.4%. It is rather compact with genic regions accounting for 87% of the total bases. This mitogenome encodes two ribosomal RNAs (rnl and rns), 25 tRNAs and 14 conserved proteins of the oxidative phosphorylation system. These tRNA genes code for all 20 standard amino acids and are arranged into three clusters. The majority of tRNA genes are clustered upstream (trnV, I, S, W, P) and downstream (trnT, E, M, M, L, A, F, K, L, Q, H, M) of the rnl gene, and a set of four tRNA genes (trnY, D, S, N) are located between rns and cox3. The 14 protein-coding genes encode NAD dehydrogenases (nad1–6 and nad4L), cytochrome oxidases (cox1–3), apocytochrome b (cob) and ATP synthases (atp6, 8, 9). For the two neighboring gene pairs, nad2/nad3 and nad4L/nad5, corresponding genes overlap by one nucleotide, which is characteristic of Cordycipitaceae (Fan et al. Citation2019). No intergenic free-standing ORF is identified. One group IA intron (designated as mL2450) interrupts rnl and contains an ORF (orf443) encoding ribosomal protein S3.
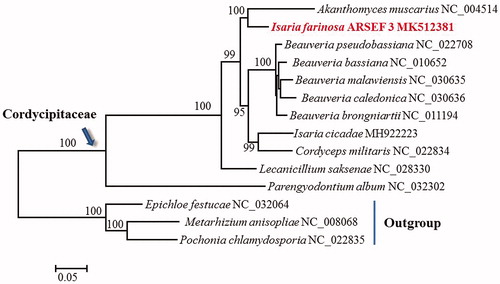
Phylogenetic analysis based on mitochondrial DNA sequences confirms I. farinosa as a member of Cordycipitaceae (). The fungus is closely related to Akanthomyces muscarius, and their clustering receives 100% support. It should be noted that I. farisona might represent a polyphyletic species complex (Dai et al. Citation2016; Kepler et al. Citation2017). The fungus shows a high genetic diversity even at a local region (Sun et al. Citation2011; Dai et al. Citation2015). Because the ex-epitype isolate (CBS 111113) of I. farinosa (type species of Isaria) clustered in the Cordyceps clade, Isaria is proposed to be rejected in favor of Cordyceps, and thus CBS 111113 has the new name Cordyceps farinosa (Kepler et al. Citation2017). Two other I. farinosa isolates, CBS 240.32 and 262.58, that are genetically distant to CBS 111113 are recently re-named as Samsoniella alboaurantium (Mongkolsamrit et al. Citation2018). Currently, there are a total of 334 I. farinosa records only in the ARSEF catalogue. The taxonomy of I. farinosa isolates (including ARSEF 3) deposited in public culture collections needs to be reappraised.
Figure 1. Phylogenetic analysis of Cordycipitaceae species based on 13 mitochondrial protein-encoding genes. Concatenated nucleotide sequences of atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, and nad6, a total of 10,272 characters, were used. The gene nad5 was excluded due to its significant phylogenetic conflict with cox1 and nad3. Three Clavicipitaceae species (Epichloe festucae, Metarhizium anisopliae, and Pochonia chlamydosporia) were used as outgroups. The tree was constructed using the maximum likelihood approach as implemented in RAxML. Support values were given for nodes that received ≥70% bootstrap values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Dai Y, Yu H, Chen Z, Yang J. 2015. Genetic differentiation of an entomogenous fungus Isaria farinosa in Yunnan, China. Mycosystema. 34:38–52.
- Dai Y, Yu H, Zeng W, Yang J, He L. 2016. Multilocus phylogenetic analyses of the genus Isaria (Ascomycota, Cordycipitaceae). Mygosystema. 35:147–160.
- Fan W-W, Zhang S, Zhang Y-J. 2019. The complete mitochondrial genome of the Chan-hua fungus Isaria cicadae: a tale of intron evolution in Cordycipitaceae. Environ Microbiol Microbiol. 21:864–879.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kepler RM, Luangsa-Ard JJ, Hywel-Jones NL, Quandt CA, Sung G-H, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, et al. 2017. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus. 8:335.
- Mongkolsamrit S, Noisripoom W, Thanakitpipattana D, Wutikhun T, Spatafora JW, Luangsa-Ard J. 2018. Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia. 110:230–257.
- Pacioni G, Frizzi G. 1978. Paecilomyces farinosus, the conidial state of Cordyceps memorabilis. Can J Bot. 56:391–394.
- Sun Z-H, Luan F-G, Zhang D-M, Chen M-J, Wang B, Li Z-Z. 2011. Genetic differentiation of Isaria farinosa populations in Anhui Province of East China. Chinese J Appl Ecol. 22:3039–3046.
- Zhang Y-J, Yang X-Q, Zhang S, Humber RA, Xu J. 2017. Genomic analyses reveal low mitochondrial and high nuclear diversity in the cyclosporin-producing fungus Tolypocladium inflatum. Appl Microbiol Biotechnol. 101:8517–8531.
- Zimmermann G. 2008. The entomopathogenic fungi Isaria farinosa (formerly Paecilomyces farinosus) and the Isaria fumosorosea species complex (formerly Paecilomyces fumosoroseus): biology, ecology and use in biological control. Biocontrol Sci Technol. 18:865–901.
